Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
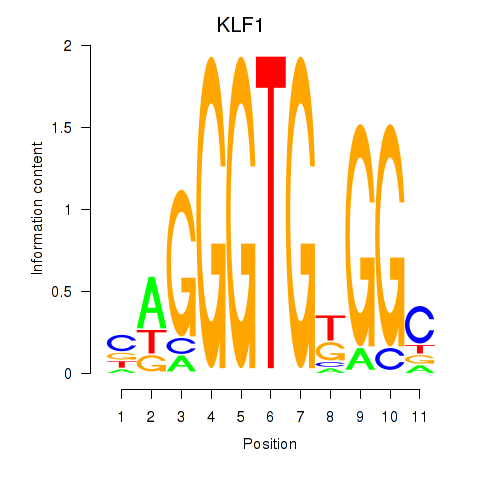
Results for KLF1
Z-value: 0.83
Transcription factors associated with KLF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF1
|
ENSG00000105610.4 | Kruppel like factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF1 | hg19_v2_chr19_-_12997995_12998021 | 0.40 | 4.8e-02 | Click! |
Activity profile of KLF1 motif
Sorted Z-values of KLF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_101963482 | 1.70 |
ENST00000419477.2
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr17_+_7210898 | 1.62 |
ENST00000572815.1
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr22_-_39639021 | 1.54 |
ENST00000455790.1
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr5_-_115910630 | 1.27 |
ENST00000343348.6
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr5_-_131826457 | 1.23 |
ENST00000437654.1
ENST00000245414.4 |
IRF1
|
interferon regulatory factor 1 |
| chr3_-_127542021 | 1.16 |
ENST00000434178.2
|
MGLL
|
monoglyceride lipase |
| chr9_+_35732312 | 1.12 |
ENST00000353704.2
|
CREB3
|
cAMP responsive element binding protein 3 |
| chr15_+_90931450 | 1.07 |
ENST00000268182.5
ENST00000560738.1 ENST00000560418.1 |
IQGAP1
|
IQ motif containing GTPase activating protein 1 |
| chrX_-_71526741 | 1.06 |
ENST00000454225.1
|
CITED1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr10_+_104155450 | 1.06 |
ENST00000471698.1
ENST00000189444.6 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr7_+_134430212 | 1.04 |
ENST00000436461.2
|
CALD1
|
caldesmon 1 |
| chr5_-_115910091 | 0.96 |
ENST00000257414.8
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr16_-_31161380 | 0.95 |
ENST00000569305.1
ENST00000418068.2 ENST00000268281.4 |
PRSS36
|
protease, serine, 36 |
| chr11_-_73471655 | 0.95 |
ENST00000400470.2
|
RAB6A
|
RAB6A, member RAS oncogene family |
| chr16_+_50776021 | 0.94 |
ENST00000566679.2
ENST00000564634.1 ENST00000398568.2 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr9_+_74764278 | 0.94 |
ENST00000238018.4
ENST00000376989.3 |
GDA
|
guanine deaminase |
| chr8_-_101962777 | 0.89 |
ENST00000395951.3
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr6_-_3227877 | 0.87 |
ENST00000259818.7
|
TUBB2B
|
tubulin, beta 2B class IIb |
| chr18_-_48723690 | 0.84 |
ENST00000406189.3
|
MEX3C
|
mex-3 RNA binding family member C |
| chr19_+_8117636 | 0.84 |
ENST00000253451.4
ENST00000315626.4 |
CCL25
|
chemokine (C-C motif) ligand 25 |
| chr5_-_95297534 | 0.82 |
ENST00000513343.1
ENST00000431061.2 |
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr11_+_66384053 | 0.79 |
ENST00000310137.4
ENST00000393979.3 ENST00000409372.1 ENST00000443702.1 ENST00000409738.4 ENST00000514361.3 ENST00000503028.2 ENST00000412278.2 ENST00000500635.2 |
RBM14
RBM4
RBM14-RBM4
|
RNA binding motif protein 14 RNA binding motif protein 4 RBM14-RBM4 readthrough |
| chr6_-_43596899 | 0.78 |
ENST00000307126.5
ENST00000452781.1 |
GTPBP2
|
GTP binding protein 2 |
| chr12_-_57504069 | 0.78 |
ENST00000543873.2
ENST00000554663.1 ENST00000557635.1 |
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr16_+_50775948 | 0.76 |
ENST00000569681.1
ENST00000569418.1 ENST00000540145.1 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr2_-_153573965 | 0.76 |
ENST00000448428.1
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr17_+_7210921 | 0.74 |
ENST00000573542.1
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr1_-_27816556 | 0.73 |
ENST00000536657.1
|
WASF2
|
WAS protein family, member 2 |
| chr3_-_186080012 | 0.72 |
ENST00000544847.1
ENST00000265022.3 |
DGKG
|
diacylglycerol kinase, gamma 90kDa |
| chr9_-_38069208 | 0.70 |
ENST00000377707.3
ENST00000377700.4 |
SHB
|
Src homology 2 domain containing adaptor protein B |
| chr16_+_50775971 | 0.70 |
ENST00000311559.9
ENST00000564326.1 ENST00000566206.1 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr7_+_86274145 | 0.67 |
ENST00000439827.1
ENST00000394720.2 ENST00000421579.1 |
GRM3
|
glutamate receptor, metabotropic 3 |
| chr10_+_120967072 | 0.67 |
ENST00000392870.2
|
GRK5
|
G protein-coupled receptor kinase 5 |
| chr5_-_95297678 | 0.66 |
ENST00000237853.4
|
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr17_-_39507064 | 0.65 |
ENST00000007735.3
|
KRT33A
|
keratin 33A |
| chr1_+_183441500 | 0.64 |
ENST00000456731.2
|
SMG7
|
SMG7 nonsense mediated mRNA decay factor |
| chr8_+_123793633 | 0.64 |
ENST00000314393.4
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr22_-_36357671 | 0.63 |
ENST00000408983.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr2_-_160919112 | 0.63 |
ENST00000283243.7
ENST00000392771.1 |
PLA2R1
|
phospholipase A2 receptor 1, 180kDa |
| chr17_-_1619491 | 0.62 |
ENST00000570416.1
ENST00000575626.1 ENST00000610106.1 ENST00000608198.1 ENST00000609442.1 ENST00000334146.3 ENST00000576489.1 ENST00000608245.1 ENST00000609398.1 ENST00000608913.1 ENST00000574016.1 ENST00000571091.1 ENST00000573127.1 ENST00000609990.1 ENST00000576749.1 |
MIR22HG
|
MIR22 host gene (non-protein coding) |
| chr6_+_106546808 | 0.62 |
ENST00000369089.3
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr4_+_2845655 | 0.61 |
ENST00000511797.1
ENST00000513328.2 ENST00000508277.1 ENST00000503455.2 |
ADD1
|
adducin 1 (alpha) |
| chr1_-_159893507 | 0.60 |
ENST00000368096.1
|
TAGLN2
|
transgelin 2 |
| chr17_-_1619535 | 0.59 |
ENST00000573075.1
ENST00000574306.1 |
MIR22HG
|
MIR22 host gene (non-protein coding) |
| chr12_+_100661156 | 0.58 |
ENST00000360820.2
|
SCYL2
|
SCY1-like 2 (S. cerevisiae) |
| chr15_+_100106126 | 0.58 |
ENST00000558812.1
ENST00000338042.6 |
MEF2A
|
myocyte enhancer factor 2A |
| chr8_+_120428546 | 0.57 |
ENST00000259526.3
|
NOV
|
nephroblastoma overexpressed |
| chr15_+_100106155 | 0.57 |
ENST00000557785.1
ENST00000558049.1 ENST00000449277.2 |
MEF2A
|
myocyte enhancer factor 2A |
| chr3_-_49449350 | 0.57 |
ENST00000454011.2
ENST00000445425.1 ENST00000422781.1 |
RHOA
|
ras homolog family member A |
| chr6_-_30654977 | 0.56 |
ENST00000399199.3
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18 |
| chr1_-_111743285 | 0.56 |
ENST00000357640.4
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr3_-_113464906 | 0.56 |
ENST00000477813.1
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr19_+_8117881 | 0.56 |
ENST00000390669.3
|
CCL25
|
chemokine (C-C motif) ligand 25 |
| chr17_-_7493390 | 0.55 |
ENST00000538513.2
ENST00000570788.1 ENST00000250055.2 |
SOX15
|
SRY (sex determining region Y)-box 15 |
| chr1_-_31712401 | 0.55 |
ENST00000373736.2
|
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr6_-_82462425 | 0.55 |
ENST00000369754.3
ENST00000320172.6 ENST00000369756.3 |
FAM46A
|
family with sequence similarity 46, member A |
| chr15_+_75074410 | 0.54 |
ENST00000439220.2
|
CSK
|
c-src tyrosine kinase |
| chr1_-_27816641 | 0.54 |
ENST00000430629.2
|
WASF2
|
WAS protein family, member 2 |
| chr6_-_83903600 | 0.54 |
ENST00000506587.1
ENST00000507554.1 |
PGM3
|
phosphoglucomutase 3 |
| chr22_+_38071615 | 0.53 |
ENST00000215909.5
|
LGALS1
|
lectin, galactoside-binding, soluble, 1 |
| chr3_-_88108212 | 0.52 |
ENST00000482016.1
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr17_-_42277203 | 0.52 |
ENST00000587097.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr4_-_103748696 | 0.51 |
ENST00000321805.7
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr14_+_23846210 | 0.51 |
ENST00000339180.4
ENST00000342473.4 ENST00000397227.3 ENST00000555731.1 |
CMTM5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr17_+_65374075 | 0.51 |
ENST00000581322.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr14_+_23845995 | 0.51 |
ENST00000359320.3
|
CMTM5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr1_-_167883327 | 0.51 |
ENST00000476818.2
ENST00000367851.4 ENST00000367848.1 |
ADCY10
|
adenylate cyclase 10 (soluble) |
| chrX_+_70503433 | 0.50 |
ENST00000276079.8
ENST00000373856.3 ENST00000373841.1 ENST00000420903.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr19_+_55795493 | 0.50 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr2_-_153573887 | 0.50 |
ENST00000493468.2
ENST00000545856.1 |
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr3_+_39093481 | 0.50 |
ENST00000302313.5
ENST00000544962.1 ENST00000396258.3 ENST00000418020.1 |
WDR48
|
WD repeat domain 48 |
| chr3_+_50273625 | 0.50 |
ENST00000536647.1
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
| chr9_-_34048873 | 0.49 |
ENST00000449054.1
ENST00000379239.4 ENST00000539807.1 ENST00000379238.1 ENST00000418786.2 ENST00000360802.1 ENST00000412543.1 |
UBAP2
|
ubiquitin associated protein 2 |
| chr12_-_49393092 | 0.49 |
ENST00000421952.2
|
DDN
|
dendrin |
| chr6_-_114664180 | 0.48 |
ENST00000312719.5
|
HS3ST5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5 |
| chr17_-_2614927 | 0.48 |
ENST00000435359.1
|
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr5_+_173315283 | 0.48 |
ENST00000265085.5
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr12_+_116997186 | 0.47 |
ENST00000306985.4
|
MAP1LC3B2
|
microtubule-associated protein 1 light chain 3 beta 2 |
| chr1_+_35225339 | 0.47 |
ENST00000339480.1
|
GJB4
|
gap junction protein, beta 4, 30.3kDa |
| chr8_+_38614807 | 0.46 |
ENST00000330691.6
ENST00000348567.4 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr1_-_243326612 | 0.46 |
ENST00000492145.1
ENST00000490813.1 ENST00000464936.1 |
CEP170
|
centrosomal protein 170kDa |
| chr14_+_42077552 | 0.45 |
ENST00000554120.1
|
LRFN5
|
leucine rich repeat and fibronectin type III domain containing 5 |
| chr6_-_83902933 | 0.44 |
ENST00000512866.1
ENST00000510258.1 ENST00000503094.1 ENST00000283977.4 ENST00000513973.1 ENST00000508748.1 |
PGM3
|
phosphoglucomutase 3 |
| chr7_-_25164868 | 0.44 |
ENST00000409409.1
ENST00000409764.1 ENST00000413447.1 |
CYCS
|
cytochrome c, somatic |
| chr7_-_80548667 | 0.44 |
ENST00000265361.3
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr15_-_64648273 | 0.43 |
ENST00000607537.1
ENST00000303052.7 ENST00000303032.6 |
CSNK1G1
|
casein kinase 1, gamma 1 |
| chr19_-_18654293 | 0.43 |
ENST00000597547.1
ENST00000222308.4 ENST00000544835.3 ENST00000610101.1 ENST00000597960.3 ENST00000608443.1 |
FKBP8
|
FK506 binding protein 8, 38kDa |
| chr17_-_7141490 | 0.42 |
ENST00000574236.1
ENST00000572789.1 |
PHF23
|
PHD finger protein 23 |
| chr4_-_10117949 | 0.41 |
ENST00000508079.1
|
WDR1
|
WD repeat domain 1 |
| chr2_+_102314161 | 0.41 |
ENST00000425019.1
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr1_-_204183071 | 0.40 |
ENST00000308302.3
|
GOLT1A
|
golgi transport 1A |
| chr17_+_7210852 | 0.40 |
ENST00000576930.1
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr11_-_62521614 | 0.40 |
ENST00000527994.1
ENST00000394807.3 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chr8_-_145018080 | 0.40 |
ENST00000354589.3
|
PLEC
|
plectin |
| chr8_+_81398444 | 0.40 |
ENST00000455036.3
ENST00000426744.2 |
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr20_+_35974532 | 0.39 |
ENST00000373578.2
|
SRC
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr11_+_67183141 | 0.39 |
ENST00000531040.1
ENST00000307823.3 ENST00000423745.2 |
CARNS1
|
carnosine synthase 1 |
| chr6_-_138428613 | 0.39 |
ENST00000421351.3
|
PERP
|
PERP, TP53 apoptosis effector |
| chr6_-_131291572 | 0.39 |
ENST00000529208.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr13_+_24553933 | 0.38 |
ENST00000424834.2
ENST00000439928.2 |
SPATA13
RP11-309I15.1
|
spermatogenesis associated 13 RP11-309I15.1 |
| chr13_+_88324870 | 0.38 |
ENST00000325089.6
|
SLITRK5
|
SLIT and NTRK-like family, member 5 |
| chr19_+_8455200 | 0.38 |
ENST00000601897.1
ENST00000594216.1 |
RAB11B
|
RAB11B, member RAS oncogene family |
| chr19_+_41698927 | 0.38 |
ENST00000310054.4
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr11_+_1074870 | 0.37 |
ENST00000441003.2
ENST00000359061.5 |
MUC2
|
mucin 2, oligomeric mucus/gel-forming |
| chr12_+_7072354 | 0.37 |
ENST00000537269.1
|
U47924.27
|
U47924.27 |
| chr10_+_31610064 | 0.37 |
ENST00000446923.2
ENST00000559476.1 |
ZEB1
|
zinc finger E-box binding homeobox 1 |
| chr1_-_222885770 | 0.37 |
ENST00000355727.2
ENST00000340020.6 |
AIDA
|
axin interactor, dorsalization associated |
| chr16_-_4466622 | 0.37 |
ENST00000570645.1
ENST00000574025.1 ENST00000572898.1 ENST00000537233.2 ENST00000571059.1 ENST00000251166.4 |
CORO7
|
coronin 7 |
| chr11_-_65625330 | 0.36 |
ENST00000531407.1
|
CFL1
|
cofilin 1 (non-muscle) |
| chr16_+_30675654 | 0.36 |
ENST00000287468.5
ENST00000395073.2 |
FBRS
|
fibrosin |
| chr1_-_159894319 | 0.36 |
ENST00000320307.4
|
TAGLN2
|
transgelin 2 |
| chr3_-_197024965 | 0.36 |
ENST00000392382.2
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr3_-_197024394 | 0.36 |
ENST00000434148.1
ENST00000412364.2 ENST00000436682.1 ENST00000456699.2 ENST00000392380.2 |
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr1_-_167883353 | 0.35 |
ENST00000545172.1
|
ADCY10
|
adenylate cyclase 10 (soluble) |
| chr16_+_89989687 | 0.35 |
ENST00000315491.7
ENST00000555576.1 ENST00000554336.1 ENST00000553967.1 |
TUBB3
|
Tubulin beta-3 chain |
| chr3_-_49449521 | 0.35 |
ENST00000431929.1
ENST00000418115.1 |
RHOA
|
ras homolog family member A |
| chrX_-_48768913 | 0.35 |
ENST00000376529.3
ENST00000247138.5 ENST00000413561.2 ENST00000452555.2 ENST00000445167.2 ENST00000376512.1 |
SLC35A2
|
solute carrier family 35 (UDP-galactose transporter), member A2 |
| chr19_+_36119975 | 0.35 |
ENST00000589559.1
ENST00000360475.4 |
RBM42
|
RNA binding motif protein 42 |
| chr6_-_39282329 | 0.35 |
ENST00000373231.4
|
KCNK17
|
potassium channel, subfamily K, member 17 |
| chr9_-_37465396 | 0.34 |
ENST00000307750.4
|
ZBTB5
|
zinc finger and BTB domain containing 5 |
| chr6_+_83903061 | 0.34 |
ENST00000369724.4
ENST00000539997.1 |
RWDD2A
|
RWD domain containing 2A |
| chr10_+_98592009 | 0.34 |
ENST00000540664.1
ENST00000371103.3 |
LCOR
|
ligand dependent nuclear receptor corepressor |
| chr8_-_27115931 | 0.34 |
ENST00000523048.1
|
STMN4
|
stathmin-like 4 |
| chr20_+_60878005 | 0.34 |
ENST00000253003.2
|
ADRM1
|
adhesion regulating molecule 1 |
| chr2_-_153574480 | 0.34 |
ENST00000410080.1
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr17_-_27949911 | 0.34 |
ENST00000492276.2
ENST00000345068.5 ENST00000584602.1 |
CORO6
|
coronin 6 |
| chr5_+_149546334 | 0.34 |
ENST00000231656.8
|
CDX1
|
caudal type homeobox 1 |
| chr19_+_36120009 | 0.34 |
ENST00000589871.1
|
RBM42
|
RNA binding motif protein 42 |
| chr11_+_67183557 | 0.33 |
ENST00000445895.2
|
CARNS1
|
carnosine synthase 1 |
| chr12_-_54019755 | 0.33 |
ENST00000588078.1
ENST00000551480.2 ENST00000548118.2 ENST00000456903.4 ENST00000591834.1 |
ATF7
RP11-793H13.10
|
activating transcription factor 7 Uncharacterized protein |
| chr12_+_51632600 | 0.33 |
ENST00000549555.1
ENST00000439799.2 ENST00000425012.2 |
DAZAP2
|
DAZ associated protein 2 |
| chr12_+_51632638 | 0.33 |
ENST00000549732.2
|
DAZAP2
|
DAZ associated protein 2 |
| chr19_+_45681997 | 0.33 |
ENST00000433642.2
|
BLOC1S3
|
biogenesis of lysosomal organelles complex-1, subunit 3 |
| chr15_-_89878025 | 0.33 |
ENST00000268124.5
ENST00000442287.2 |
POLG
|
polymerase (DNA directed), gamma |
| chr7_-_138794081 | 0.32 |
ENST00000464606.1
|
ZC3HAV1
|
zinc finger CCCH-type, antiviral 1 |
| chr20_-_4804244 | 0.32 |
ENST00000379400.3
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2 |
| chr12_+_54332535 | 0.31 |
ENST00000243056.3
|
HOXC13
|
homeobox C13 |
| chr1_+_29213678 | 0.31 |
ENST00000347529.3
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr19_+_35629702 | 0.31 |
ENST00000351325.4
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr6_-_39282221 | 0.31 |
ENST00000453413.2
|
KCNK17
|
potassium channel, subfamily K, member 17 |
| chrX_-_23761317 | 0.31 |
ENST00000492081.1
ENST00000379303.5 ENST00000336430.7 |
ACOT9
|
acyl-CoA thioesterase 9 |
| chr15_+_75074385 | 0.31 |
ENST00000220003.9
|
CSK
|
c-src tyrosine kinase |
| chr11_-_57089671 | 0.31 |
ENST00000532437.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr6_-_84140757 | 0.31 |
ENST00000541327.1
ENST00000369705.3 ENST00000543031.1 |
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr19_+_46850320 | 0.30 |
ENST00000391919.1
|
PPP5C
|
protein phosphatase 5, catalytic subunit |
| chr19_+_46850251 | 0.30 |
ENST00000012443.4
|
PPP5C
|
protein phosphatase 5, catalytic subunit |
| chr19_+_35630022 | 0.30 |
ENST00000589209.1
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr11_-_62313090 | 0.30 |
ENST00000528508.1
ENST00000533365.1 |
AHNAK
|
AHNAK nucleoprotein |
| chr5_+_159895275 | 0.30 |
ENST00000517927.1
|
MIR146A
|
microRNA 146a |
| chr17_-_39526052 | 0.30 |
ENST00000251646.3
|
KRT33B
|
keratin 33B |
| chr11_-_75379612 | 0.29 |
ENST00000526740.1
|
MAP6
|
microtubule-associated protein 6 |
| chr4_+_39699664 | 0.29 |
ENST00000261427.5
ENST00000510934.1 ENST00000295963.6 |
UBE2K
|
ubiquitin-conjugating enzyme E2K |
| chrX_+_23352133 | 0.29 |
ENST00000379361.4
|
PTCHD1
|
patched domain containing 1 |
| chr12_-_54020107 | 0.29 |
ENST00000588232.1
ENST00000548446.2 ENST00000420353.2 ENST00000591397.1 ENST00000415113.1 |
ATF7
|
activating transcription factor 7 |
| chr3_+_38206975 | 0.29 |
ENST00000446845.1
ENST00000311806.3 |
OXSR1
|
oxidative stress responsive 1 |
| chr19_-_44008863 | 0.29 |
ENST00000601646.1
|
PHLDB3
|
pleckstrin homology-like domain, family B, member 3 |
| chr4_+_146403912 | 0.29 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr6_-_33548006 | 0.28 |
ENST00000374467.3
|
BAK1
|
BCL2-antagonist/killer 1 |
| chr6_-_30709980 | 0.28 |
ENST00000416018.1
ENST00000445853.1 ENST00000413165.1 ENST00000418160.1 |
FLOT1
|
flotillin 1 |
| chr17_+_30814707 | 0.28 |
ENST00000584792.1
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr1_-_23670752 | 0.28 |
ENST00000302271.6
ENST00000426846.2 ENST00000427764.2 ENST00000606561.1 ENST00000374616.3 |
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr1_+_6845384 | 0.28 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr11_+_64009072 | 0.28 |
ENST00000535135.1
ENST00000394540.3 |
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr2_+_220379052 | 0.28 |
ENST00000347842.3
ENST00000358078.4 |
ASIC4
|
acid-sensing (proton-gated) ion channel family member 4 |
| chr17_-_7590745 | 0.27 |
ENST00000514944.1
ENST00000503591.1 ENST00000455263.2 ENST00000420246.2 ENST00000445888.2 ENST00000509690.1 ENST00000604348.1 ENST00000269305.4 |
TP53
|
tumor protein p53 |
| chr16_+_67022633 | 0.27 |
ENST00000398354.1
ENST00000326686.5 |
CES4A
|
carboxylesterase 4A |
| chr5_+_71014990 | 0.27 |
ENST00000296777.4
|
CARTPT
|
CART prepropeptide |
| chr12_-_49365501 | 0.26 |
ENST00000403957.1
ENST00000301061.4 |
WNT10B
|
wingless-type MMTV integration site family, member 10B |
| chr17_+_46125707 | 0.26 |
ENST00000584137.1
ENST00000362042.3 ENST00000585291.1 ENST00000357480.5 |
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr9_-_35732362 | 0.26 |
ENST00000314888.9
ENST00000540444.1 |
TLN1
|
talin 1 |
| chr5_-_112257914 | 0.26 |
ENST00000513339.1
ENST00000545426.1 ENST00000504247.1 |
REEP5
|
receptor accessory protein 5 |
| chr19_-_10047219 | 0.26 |
ENST00000264833.4
|
OLFM2
|
olfactomedin 2 |
| chr6_+_106534192 | 0.26 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr6_-_33547975 | 0.26 |
ENST00000442998.2
ENST00000360661.5 |
BAK1
|
BCL2-antagonist/killer 1 |
| chr2_+_85360499 | 0.26 |
ENST00000282111.3
|
TCF7L1
|
transcription factor 7-like 1 (T-cell specific, HMG-box) |
| chr12_-_56694142 | 0.26 |
ENST00000550655.1
ENST00000548567.1 ENST00000551430.2 ENST00000351328.3 |
CS
|
citrate synthase |
| chr4_+_2845547 | 0.26 |
ENST00000264758.7
ENST00000446856.1 ENST00000398125.1 |
ADD1
|
adducin 1 (alpha) |
| chr8_+_81397876 | 0.25 |
ENST00000430430.1
|
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr12_-_93835665 | 0.25 |
ENST00000552442.1
ENST00000550657.1 |
UBE2N
|
ubiquitin-conjugating enzyme E2N |
| chrX_-_69479654 | 0.25 |
ENST00000374519.2
|
P2RY4
|
pyrimidinergic receptor P2Y, G-protein coupled, 4 |
| chr6_+_11094266 | 0.25 |
ENST00000416247.2
|
SMIM13
|
small integral membrane protein 13 |
| chr15_-_101142401 | 0.25 |
ENST00000314742.8
|
LINS
|
lines homolog (Drosophila) |
| chr16_+_67022582 | 0.25 |
ENST00000541479.1
ENST00000338718.4 |
CES4A
|
carboxylesterase 4A |
| chr17_+_7210294 | 0.25 |
ENST00000336452.7
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr5_+_71403061 | 0.25 |
ENST00000512974.1
ENST00000296755.7 |
MAP1B
|
microtubule-associated protein 1B |
| chr1_-_36851489 | 0.25 |
ENST00000373130.3
ENST00000373132.3 |
STK40
|
serine/threonine kinase 40 |
| chr7_+_87257854 | 0.25 |
ENST00000394654.3
|
RUNDC3B
|
RUN domain containing 3B |
| chr16_+_67022476 | 0.24 |
ENST00000540947.2
|
CES4A
|
carboxylesterase 4A |
| chrX_+_133507389 | 0.24 |
ENST00000370800.4
|
PHF6
|
PHD finger protein 6 |
| chr3_+_8775466 | 0.24 |
ENST00000343849.2
ENST00000397368.2 |
CAV3
|
caveolin 3 |
| chr8_-_101963677 | 0.24 |
ENST00000395956.3
ENST00000395953.2 |
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr1_-_23670813 | 0.24 |
ENST00000374612.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr5_-_139283982 | 0.24 |
ENST00000340391.3
|
NRG2
|
neuregulin 2 |
| chr1_-_36851475 | 0.23 |
ENST00000373129.3
|
STK40
|
serine/threonine kinase 40 |
| chr5_-_83680603 | 0.23 |
ENST00000296591.5
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3 |
| chr10_+_96443378 | 0.23 |
ENST00000285979.6
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18 |
| chr19_-_44285401 | 0.23 |
ENST00000262888.3
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr19_+_39687596 | 0.23 |
ENST00000339852.4
|
NCCRP1
|
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
| chr5_-_168006591 | 0.23 |
ENST00000239231.6
|
PANK3
|
pantothenate kinase 3 |
| chr17_+_39845134 | 0.23 |
ENST00000591776.1
ENST00000469257.1 |
EIF1
|
eukaryotic translation initiation factor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.4 | 1.5 | GO:0090678 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.4 | 3.0 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.3 | 1.2 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.3 | 1.1 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.2 | 1.5 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.2 | 1.4 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.2 | 0.9 | GO:1990869 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.2 | 0.9 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.2 | 1.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.2 | 1.0 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.2 | 1.0 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.2 | 0.8 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.2 | 0.9 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.2 | 2.8 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.2 | 0.5 | GO:0048627 | myoblast development(GO:0048627) |
| 0.2 | 0.5 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.2 | 0.5 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.2 | 0.7 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.2 | 0.9 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.2 | 0.2 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.2 | 1.2 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.2 | 1.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) ERK5 cascade(GO:0070375) |
| 0.2 | 0.6 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 2.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.9 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.4 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.1 | 0.1 | GO:1903179 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.1 | 0.4 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.1 | 0.3 | GO:2000308 | iron assimilation(GO:0033212) iron assimilation by chelation and transport(GO:0033214) positive regulation of bone mineralization involved in bone maturation(GO:1900159) negative regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000308) |
| 0.1 | 0.9 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.1 | 0.8 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.1 | 0.6 | GO:0010734 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.1 | 0.4 | GO:0016334 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.1 | 0.4 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 0.6 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.4 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.1 | 0.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.3 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 0.3 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.1 | 0.7 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.1 | 0.3 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.1 | 0.9 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.1 | 0.4 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.5 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 0.6 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 1.1 | GO:1900086 | regulation of peptidyl-tyrosine autophosphorylation(GO:1900084) positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.1 | 0.6 | GO:2000784 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.1 | 0.8 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 0.2 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.1 | 0.2 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 0.2 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 0.7 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.7 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 0.7 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.1 | 0.6 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.5 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.1 | 0.2 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.1 | 0.2 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.1 | 0.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.3 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 0.3 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.1 | 0.8 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.2 | GO:1902868 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.1 | 0.3 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 0.3 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 0.2 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.5 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.6 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 1.2 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.4 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.7 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.3 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.0 | 0.6 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.8 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.2 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.3 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.2 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.4 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.4 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.2 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.0 | 0.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.3 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.2 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.5 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.3 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.5 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.2 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.3 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.2 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.2 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.4 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.7 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.2 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 1.0 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.5 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.2 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.6 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.7 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.7 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.1 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.0 | 0.4 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.5 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.3 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.4 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.9 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.3 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.3 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.8 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.3 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.1 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.1 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.4 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.2 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.0 | 0.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.0 | GO:1902463 | establishment of Sertoli cell barrier(GO:0097368) protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.1 | GO:0090650 | rRNA transport(GO:0051029) response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 1.5 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.3 | GO:0021756 | striatum development(GO:0021756) |
| 0.0 | 0.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.2 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.0 | 0.2 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.0 | 0.4 | GO:0021904 | dorsal/ventral neural tube patterning(GO:0021904) |
| 0.0 | 0.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.0 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.0 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.1 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.2 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.0 | 0.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.5 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.3 | 3.0 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.2 | 0.9 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 1.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.8 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.6 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.3 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.1 | 0.3 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.4 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.1 | 1.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.7 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 1.6 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 0.2 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.1 | 0.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.3 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 0.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.3 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.0 | 0.2 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 1.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 1.8 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 1.0 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.6 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 1.3 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 3.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 2.9 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 2.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.3 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.0 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.3 | 0.8 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.2 | 0.7 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.2 | 2.6 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.2 | 0.6 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.2 | 3.0 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.2 | 0.9 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 1.0 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.7 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.5 | GO:0030395 | lactose binding(GO:0030395) |
| 0.1 | 0.4 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 1.5 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 1.5 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 0.3 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.2 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.1 | 0.2 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.1 | 0.2 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 0.7 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 1.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 1.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 1.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.5 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.1 | 0.5 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.5 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.8 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.2 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.1 | 0.3 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 1.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.4 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 1.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 2.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.3 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.3 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.9 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.5 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.3 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.4 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 1.0 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.1 | GO:0005220 | inositol hexakisphosphate binding(GO:0000822) inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.7 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.5 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.6 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.1 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.0 | 0.5 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 3.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0097001 | glycosphingolipid binding(GO:0043208) ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.6 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.3 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 1.5 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.1 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.0 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.0 | 1.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.8 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 3.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 2.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 3.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.7 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.8 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 1.9 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 1.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.2 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.7 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.0 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.1 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.2 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 1.1 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 3.0 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 1.0 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 3.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.1 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 1.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.0 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.9 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 1.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.3 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.4 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 1.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.6 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.8 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.4 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |