Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
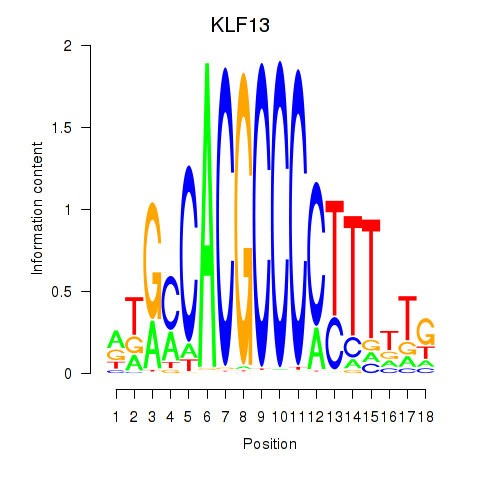
Results for KLF13
Z-value: 0.58
Transcription factors associated with KLF13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF13
|
ENSG00000169926.5 | Kruppel like factor 13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF13 | hg19_v2_chr15_+_31619013_31619095 | -0.21 | 3.0e-01 | Click! |
Activity profile of KLF13 motif
Sorted Z-values of KLF13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_82641706 | 1.17 |
ENST00000439287.4
|
GOLGA6L10
|
golgin A6 family-like 10 |
| chr19_+_45504688 | 1.07 |
ENST00000221452.8
ENST00000540120.1 ENST00000505236.1 |
RELB
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr8_-_37756972 | 0.90 |
ENST00000330843.4
ENST00000522727.1 ENST00000287263.4 |
RAB11FIP1
|
RAB11 family interacting protein 1 (class I) |
| chr13_+_24734880 | 0.79 |
ENST00000382095.4
|
SPATA13
|
spermatogenesis associated 13 |
| chr15_+_30375158 | 0.72 |
ENST00000341650.6
ENST00000567927.1 |
GOLGA8J
|
golgin A8 family, member J |
| chr15_-_82939157 | 0.69 |
ENST00000559949.1
|
RP13-996F3.5
|
golgin A6 family-like 18 |
| chr19_-_55672037 | 0.68 |
ENST00000588076.1
|
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr7_+_86273218 | 0.65 |
ENST00000361669.2
|
GRM3
|
glutamate receptor, metabotropic 3 |
| chr15_+_30896329 | 0.61 |
ENST00000566740.1
|
GOLGA8H
|
golgin A8 family, member H |
| chr6_+_31126291 | 0.59 |
ENST00000376257.3
ENST00000376255.4 |
TCF19
|
transcription factor 19 |
| chr2_+_114737472 | 0.53 |
ENST00000420161.1
|
AC110769.3
|
AC110769.3 |
| chr12_+_112563335 | 0.47 |
ENST00000549358.1
ENST00000257604.5 ENST00000548092.1 ENST00000552896.1 |
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr12_+_112563303 | 0.46 |
ENST00000412615.2
|
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr19_+_49867181 | 0.45 |
ENST00000597546.1
|
DKKL1
|
dickkopf-like 1 |
| chr8_-_145013711 | 0.43 |
ENST00000345136.3
|
PLEC
|
plectin |
| chr11_+_66384053 | 0.42 |
ENST00000310137.4
ENST00000393979.3 ENST00000409372.1 ENST00000443702.1 ENST00000409738.4 ENST00000514361.3 ENST00000503028.2 ENST00000412278.2 ENST00000500635.2 |
RBM14
RBM4
RBM14-RBM4
|
RNA binding motif protein 14 RNA binding motif protein 4 RBM14-RBM4 readthrough |
| chr17_-_74497432 | 0.40 |
ENST00000590288.1
ENST00000313080.4 ENST00000592123.1 ENST00000591255.1 ENST00000585989.1 ENST00000591697.1 ENST00000389760.4 |
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila) |
| chr17_+_7338737 | 0.38 |
ENST00000323206.1
ENST00000396568.1 |
TMEM102
|
transmembrane protein 102 |
| chr5_-_32313019 | 0.37 |
ENST00000280285.5
ENST00000264934.5 |
MTMR12
|
myotubularin related protein 12 |
| chr7_+_5632436 | 0.37 |
ENST00000340250.6
ENST00000382361.3 |
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
| chr15_+_40453204 | 0.37 |
ENST00000287598.6
ENST00000412359.3 |
BUB1B
|
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr12_+_58120044 | 0.36 |
ENST00000542466.2
|
AGAP2-AS1
|
AGAP2 antisense RNA 1 |
| chr19_-_8213753 | 0.35 |
ENST00000601739.1
|
FBN3
|
fibrillin 3 |
| chr6_+_31588478 | 0.35 |
ENST00000376007.4
ENST00000376033.2 |
PRRC2A
|
proline-rich coiled-coil 2A |
| chr6_+_33172407 | 0.34 |
ENST00000374662.3
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr19_+_39971505 | 0.34 |
ENST00000544017.1
|
TIMM50
|
translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae) |
| chr19_+_45349630 | 0.34 |
ENST00000252483.5
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
| chr11_-_89653576 | 0.34 |
ENST00000420869.1
|
TRIM49D1
|
tripartite motif containing 49D1 |
| chrX_+_72062617 | 0.34 |
ENST00000440247.1
|
DMRTC1B
|
DMRT-like family C1B |
| chr7_-_23053719 | 0.34 |
ENST00000432176.2
ENST00000440481.1 |
FAM126A
|
family with sequence similarity 126, member A |
| chr7_-_23053693 | 0.32 |
ENST00000409763.1
ENST00000409923.1 |
FAM126A
|
family with sequence similarity 126, member A |
| chr6_-_11779174 | 0.32 |
ENST00000379413.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr19_+_39971470 | 0.32 |
ENST00000607714.1
ENST00000599794.1 ENST00000597666.1 ENST00000601403.1 ENST00000602028.1 |
TIMM50
|
translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae) |
| chr15_-_32695396 | 0.31 |
ENST00000512626.2
ENST00000435655.2 |
GOLGA8K
AC139426.2
|
golgin A8 family, member K Uncharacterized protein; cDNA FLJ52611 |
| chr6_+_41040678 | 0.31 |
ENST00000341376.6
ENST00000353205.5 |
NFYA
|
nuclear transcription factor Y, alpha |
| chr6_-_11779014 | 0.31 |
ENST00000229583.5
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr6_-_43596899 | 0.30 |
ENST00000307126.5
ENST00000452781.1 |
GTPBP2
|
GTP binding protein 2 |
| chr15_-_28957469 | 0.29 |
ENST00000563027.1
ENST00000340249.3 |
GOLGA8M
|
golgin A8 family, member M |
| chr2_-_239140011 | 0.29 |
ENST00000409376.1
ENST00000409070.1 ENST00000409942.1 |
AC016757.3
|
Protein LOC151174 |
| chr5_-_32313082 | 0.29 |
ENST00000382142.3
|
MTMR12
|
myotubularin related protein 12 |
| chr19_+_50887585 | 0.28 |
ENST00000440232.2
ENST00000601098.1 ENST00000599857.1 ENST00000593887.1 |
POLD1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr19_-_50979981 | 0.28 |
ENST00000595790.1
ENST00000600100.1 |
FAM71E1
|
family with sequence similarity 71, member E1 |
| chr15_+_52311398 | 0.27 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr6_-_11779403 | 0.27 |
ENST00000414691.3
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr17_-_77770830 | 0.27 |
ENST00000269385.4
|
CBX8
|
chromobox homolog 8 |
| chr19_-_55972936 | 0.26 |
ENST00000425675.2
ENST00000589080.1 ENST00000085068.3 |
ISOC2
|
isochorismatase domain containing 2 |
| chr19_+_45349432 | 0.26 |
ENST00000252485.4
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
| chr2_-_239140276 | 0.25 |
ENST00000334973.4
|
AC016757.3
|
Protein LOC151174 |
| chr18_-_56940611 | 0.25 |
ENST00000256852.7
ENST00000334889.3 |
RAX
|
retina and anterior neural fold homeobox |
| chr19_+_10765003 | 0.24 |
ENST00000407004.3
ENST00000589998.1 ENST00000589600.1 |
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chr19_-_36236292 | 0.24 |
ENST00000378975.3
ENST00000412391.2 ENST00000292879.5 |
U2AF1L4
|
U2 small nuclear RNA auxiliary factor 1-like 4 |
| chr17_+_19281034 | 0.24 |
ENST00000308406.5
ENST00000299612.7 |
MAPK7
|
mitogen-activated protein kinase 7 |
| chr6_-_37467628 | 0.24 |
ENST00000373408.3
|
CCDC167
|
coiled-coil domain containing 167 |
| chr4_-_10117949 | 0.23 |
ENST00000508079.1
|
WDR1
|
WD repeat domain 1 |
| chr19_+_45458503 | 0.23 |
ENST00000337392.5
ENST00000591304.1 |
CLPTM1
|
cleft lip and palate associated transmembrane protein 1 |
| chr19_+_10764937 | 0.23 |
ENST00000449870.1
ENST00000318511.3 ENST00000420083.1 |
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chr7_-_92463210 | 0.23 |
ENST00000265734.4
|
CDK6
|
cyclin-dependent kinase 6 |
| chr2_+_114737146 | 0.22 |
ENST00000412690.1
|
AC010982.1
|
AC010982.1 |
| chr2_+_239335449 | 0.21 |
ENST00000264607.4
|
ASB1
|
ankyrin repeat and SOCS box containing 1 |
| chr20_-_8000426 | 0.21 |
ENST00000527925.1
ENST00000246024.2 |
TMX4
|
thioredoxin-related transmembrane protein 4 |
| chr19_-_2151523 | 0.21 |
ENST00000350812.6
ENST00000355272.6 ENST00000356926.4 ENST00000345016.5 |
AP3D1
|
adaptor-related protein complex 3, delta 1 subunit |
| chr12_+_121124599 | 0.21 |
ENST00000228506.3
|
MLEC
|
malectin |
| chr20_+_48599506 | 0.21 |
ENST00000244050.2
|
SNAI1
|
snail family zinc finger 1 |
| chr19_+_39616410 | 0.21 |
ENST00000602004.1
ENST00000599470.1 ENST00000321944.4 ENST00000593480.1 ENST00000358301.3 ENST00000593690.1 ENST00000599386.1 |
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr20_-_44539538 | 0.21 |
ENST00000372420.1
|
PLTP
|
phospholipid transfer protein |
| chr19_+_49866851 | 0.21 |
ENST00000221498.2
ENST00000596402.1 |
DKKL1
|
dickkopf-like 1 |
| chr11_-_47574664 | 0.20 |
ENST00000310513.5
ENST00000531165.1 |
CELF1
|
CUGBP, Elav-like family member 1 |
| chr4_-_13546632 | 0.20 |
ENST00000382438.5
|
NKX3-2
|
NK3 homeobox 2 |
| chr19_-_7936344 | 0.20 |
ENST00000599142.1
|
CTD-3193O13.9
|
Protein FLJ22184 |
| chr19_+_36236491 | 0.20 |
ENST00000591949.1
|
PSENEN
|
presenilin enhancer gamma secretase subunit |
| chrX_+_47077632 | 0.19 |
ENST00000457458.2
|
CDK16
|
cyclin-dependent kinase 16 |
| chr4_+_699610 | 0.19 |
ENST00000521023.2
|
PCGF3
|
polycomb group ring finger 3 |
| chr16_-_3074231 | 0.19 |
ENST00000572355.1
ENST00000248089.3 ENST00000574980.1 ENST00000354679.3 ENST00000396916.1 ENST00000573842.1 |
HCFC1R1
|
host cell factor C1 regulator 1 (XPO1 dependent) |
| chr11_+_44587141 | 0.19 |
ENST00000227155.4
ENST00000342935.3 ENST00000532544.1 |
CD82
|
CD82 molecule |
| chr15_+_41851211 | 0.18 |
ENST00000263798.3
|
TYRO3
|
TYRO3 protein tyrosine kinase |
| chr15_-_74374891 | 0.18 |
ENST00000290438.3
|
GOLGA6A
|
golgin A6 family, member A |
| chr17_+_6915902 | 0.18 |
ENST00000570898.1
ENST00000552842.1 |
RNASEK
|
ribonuclease, RNase K |
| chr1_-_42921915 | 0.18 |
ENST00000372565.3
ENST00000433602.2 |
ZMYND12
|
zinc finger, MYND-type containing 12 |
| chr19_-_44100275 | 0.18 |
ENST00000422989.1
ENST00000598324.1 |
IRGQ
|
immunity-related GTPase family, Q |
| chr16_+_3074002 | 0.18 |
ENST00000326266.8
ENST00000574549.1 ENST00000575576.1 ENST00000253952.9 |
THOC6
|
THO complex 6 homolog (Drosophila) |
| chr19_-_3557401 | 0.18 |
ENST00000389395.3
ENST00000398558.4 |
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr2_+_71295733 | 0.18 |
ENST00000443938.2
ENST00000244204.6 |
NAGK
|
N-acetylglucosamine kinase |
| chr10_+_134351319 | 0.17 |
ENST00000368594.3
ENST00000368593.3 |
INPP5A
|
inositol polyphosphate-5-phosphatase, 40kDa |
| chr6_+_106534192 | 0.17 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr16_-_3073933 | 0.17 |
ENST00000574151.1
|
HCFC1R1
|
host cell factor C1 regulator 1 (XPO1 dependent) |
| chr15_-_74988281 | 0.17 |
ENST00000566828.1
ENST00000563009.1 ENST00000568176.1 ENST00000566243.1 ENST00000566219.1 ENST00000426797.3 ENST00000566119.1 ENST00000315127.4 |
EDC3
|
enhancer of mRNA decapping 3 |
| chr17_+_6915798 | 0.17 |
ENST00000402093.1
|
RNASEK
|
ribonuclease, RNase K |
| chr17_+_6915730 | 0.17 |
ENST00000548577.1
|
RNASEK
|
ribonuclease, RNase K |
| chr17_+_79679369 | 0.17 |
ENST00000350690.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr19_+_36236514 | 0.17 |
ENST00000222266.2
|
PSENEN
|
presenilin enhancer gamma secretase subunit |
| chr15_-_30685752 | 0.17 |
ENST00000299847.2
ENST00000397827.3 |
CHRFAM7A
|
CHRNA7 (cholinergic receptor, nicotinic, alpha 7, exons 5-10) and FAM7A (family with sequence similarity 7A, exons A-E) fusion |
| chr7_+_100271446 | 0.16 |
ENST00000419828.1
ENST00000427895.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr16_+_4526341 | 0.16 |
ENST00000458134.3
ENST00000219700.6 ENST00000575120.1 ENST00000572812.1 ENST00000574466.1 ENST00000576827.1 ENST00000570445.1 |
HMOX2
|
heme oxygenase (decycling) 2 |
| chr5_+_38258511 | 0.16 |
ENST00000354891.3
ENST00000322350.5 |
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains |
| chr20_+_34129770 | 0.16 |
ENST00000348547.2
ENST00000357394.4 ENST00000447986.1 ENST00000279052.6 ENST00000416206.1 ENST00000411577.1 ENST00000413587.1 |
ERGIC3
|
ERGIC and golgi 3 |
| chr16_-_28936493 | 0.16 |
ENST00000544477.1
ENST00000357573.6 |
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr16_-_89883015 | 0.16 |
ENST00000563673.1
ENST00000389301.3 ENST00000568369.1 ENST00000534992.1 ENST00000389302.3 ENST00000543736.1 |
FANCA
|
Fanconi anemia, complementation group A |
| chr7_+_100271355 | 0.15 |
ENST00000436220.1
ENST00000424361.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr10_-_111683183 | 0.15 |
ENST00000403138.2
ENST00000369683.1 |
XPNPEP1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr11_+_49050504 | 0.15 |
ENST00000332682.7
|
TRIM49B
|
tripartite motif containing 49B |
| chrX_-_72097698 | 0.15 |
ENST00000373530.1
|
DMRTC1
|
DMRT-like family C1 |
| chr14_-_67981916 | 0.15 |
ENST00000357461.2
|
TMEM229B
|
transmembrane protein 229B |
| chrX_+_72062802 | 0.15 |
ENST00000373533.1
|
DMRTC1B
|
DMRT-like family C1B |
| chr10_-_111683308 | 0.15 |
ENST00000502935.1
ENST00000322238.8 ENST00000369680.4 |
XPNPEP1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr7_-_99698338 | 0.15 |
ENST00000354230.3
ENST00000425308.1 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr10_+_104678102 | 0.14 |
ENST00000433628.2
|
CNNM2
|
cyclin M2 |
| chr11_-_59383617 | 0.14 |
ENST00000263847.1
|
OSBP
|
oxysterol binding protein |
| chr4_+_95373037 | 0.14 |
ENST00000359265.4
ENST00000437932.1 ENST00000380180.3 ENST00000318007.5 ENST00000450793.1 ENST00000538141.1 ENST00000317968.4 ENST00000512274.1 ENST00000503974.1 ENST00000504489.1 ENST00000542407.1 |
PDLIM5
|
PDZ and LIM domain 5 |
| chr17_+_79679299 | 0.14 |
ENST00000331531.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr2_+_71295717 | 0.14 |
ENST00000418807.3
ENST00000443872.2 |
NAGK
|
N-acetylglucosamine kinase |
| chr15_+_72947079 | 0.14 |
ENST00000421285.3
|
GOLGA6B
|
golgin A6 family, member B |
| chr10_+_101292684 | 0.14 |
ENST00000344586.7
|
NKX2-3
|
NK2 homeobox 3 |
| chr10_+_104678032 | 0.13 |
ENST00000369878.4
ENST00000369875.3 |
CNNM2
|
cyclin M2 |
| chrX_+_134555863 | 0.13 |
ENST00000417443.2
|
LINC00086
|
long intergenic non-protein coding RNA 86 |
| chr7_+_99775520 | 0.13 |
ENST00000317296.5
ENST00000422690.1 ENST00000439782.1 |
STAG3
|
stromal antigen 3 |
| chr1_-_111506562 | 0.13 |
ENST00000485275.2
ENST00000369763.4 |
LRIF1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr4_+_699537 | 0.13 |
ENST00000419774.1
ENST00000362003.5 ENST00000400151.2 ENST00000427463.1 ENST00000470161.2 |
PCGF3
|
polycomb group ring finger 3 |
| chr19_+_49866331 | 0.13 |
ENST00000597873.1
|
DKKL1
|
dickkopf-like 1 |
| chr1_+_45805728 | 0.13 |
ENST00000539779.1
|
TOE1
|
target of EGR1, member 1 (nuclear) |
| chr15_-_101817616 | 0.13 |
ENST00000526049.1
ENST00000398226.3 ENST00000537379.1 |
VIMP
|
VCP-interacting membrane protein |
| chr12_+_58013693 | 0.12 |
ENST00000320442.4
ENST00000379218.2 |
SLC26A10
|
solute carrier family 26, member 10 |
| chr7_+_99775366 | 0.12 |
ENST00000394018.2
ENST00000416412.1 |
STAG3
|
stromal antigen 3 |
| chr19_-_12792585 | 0.12 |
ENST00000351660.5
|
DHPS
|
deoxyhypusine synthase |
| chr15_-_20747114 | 0.12 |
ENST00000427390.2
|
GOLGA6L6
|
golgin A6 family-like 6 |
| chr11_-_102962929 | 0.12 |
ENST00000260247.5
|
DCUN1D5
|
DCN1, defective in cullin neddylation 1, domain containing 5 |
| chr17_-_19281203 | 0.12 |
ENST00000487415.2
|
B9D1
|
B9 protein domain 1 |
| chr1_-_11741155 | 0.12 |
ENST00000445656.1
ENST00000376669.5 ENST00000456915.1 ENST00000376692.4 |
MAD2L2
|
MAD2 mitotic arrest deficient-like 2 (yeast) |
| chr11_-_102962874 | 0.12 |
ENST00000531543.1
|
DCUN1D5
|
DCN1, defective in cullin neddylation 1, domain containing 5 |
| chr12_-_49961883 | 0.12 |
ENST00000553173.1
ENST00000550165.1 ENST00000546244.1 ENST00000343810.4 |
MCRS1
|
microspherule protein 1 |
| chr11_-_82612727 | 0.11 |
ENST00000531128.1
ENST00000535099.1 ENST00000527444.1 |
PRCP
|
prolylcarboxypeptidase (angiotensinase C) |
| chr13_-_95953589 | 0.11 |
ENST00000538287.1
ENST00000376887.4 ENST00000412704.1 ENST00000536256.1 ENST00000431522.1 |
ABCC4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr13_-_44980021 | 0.11 |
ENST00000432701.2
ENST00000607312.1 |
LINC01071
|
long intergenic non-protein coding RNA 1071 |
| chrX_-_13062752 | 0.11 |
ENST00000380625.3
ENST00000542843.1 |
FAM9C
|
family with sequence similarity 9, member C |
| chr19_+_50979753 | 0.11 |
ENST00000597426.1
ENST00000334976.6 ENST00000376918.3 ENST00000598585.1 |
EMC10
|
ER membrane protein complex subunit 10 |
| chr21_+_44394742 | 0.11 |
ENST00000432907.2
|
PKNOX1
|
PBX/knotted 1 homeobox 1 |
| chr11_-_89540388 | 0.10 |
ENST00000532501.2
|
TRIM49
|
tripartite motif containing 49 |
| chr2_-_85645545 | 0.10 |
ENST00000409275.1
|
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr11_+_65479702 | 0.10 |
ENST00000530446.1
ENST00000534104.1 ENST00000530605.1 ENST00000528198.1 ENST00000531880.1 ENST00000534650.1 |
KAT5
|
K(lysine) acetyltransferase 5 |
| chr17_-_74733404 | 0.10 |
ENST00000508921.3
ENST00000583836.1 ENST00000358156.6 ENST00000392485.2 ENST00000359995.5 |
SRSF2
|
serine/arginine-rich splicing factor 2 |
| chr12_-_58240470 | 0.10 |
ENST00000548823.1
ENST00000398073.2 |
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr12_+_105724414 | 0.09 |
ENST00000443585.1
ENST00000552457.1 ENST00000549893.1 |
C12orf75
|
chromosome 12 open reading frame 75 |
| chr19_+_5455421 | 0.09 |
ENST00000222033.4
|
ZNRF4
|
zinc and ring finger 4 |
| chr20_+_37590942 | 0.09 |
ENST00000373325.2
ENST00000252011.3 ENST00000373323.4 |
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr15_+_22736484 | 0.09 |
ENST00000560659.2
|
GOLGA6L1
|
golgin A6 family-like 1 |
| chr17_-_40333150 | 0.09 |
ENST00000264661.3
|
KCNH4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr19_-_12792704 | 0.09 |
ENST00000210060.7
|
DHPS
|
deoxyhypusine synthase |
| chr15_+_22736246 | 0.09 |
ENST00000316397.3
|
GOLGA6L1
|
golgin A6 family-like 1 |
| chrX_-_13062697 | 0.09 |
ENST00000333995.3
|
FAM9C
|
family with sequence similarity 9, member C |
| chr15_-_75017711 | 0.09 |
ENST00000567032.1
ENST00000564596.1 ENST00000566503.1 ENST00000395049.4 ENST00000395048.2 ENST00000379727.3 |
CYP1A1
|
cytochrome P450, family 1, subfamily A, polypeptide 1 |
| chr17_-_13505219 | 0.08 |
ENST00000284110.1
|
HS3ST3A1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
| chr19_-_6767516 | 0.08 |
ENST00000245908.6
|
SH2D3A
|
SH2 domain containing 3A |
| chr10_+_104005272 | 0.08 |
ENST00000369983.3
|
GBF1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr19_-_6767431 | 0.08 |
ENST00000437152.3
ENST00000597687.1 |
SH2D3A
|
SH2 domain containing 3A |
| chr19_-_47164386 | 0.08 |
ENST00000391916.2
ENST00000410105.2 |
DACT3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr19_-_50370509 | 0.08 |
ENST00000596014.1
|
PNKP
|
polynucleotide kinase 3'-phosphatase |
| chr14_+_68286516 | 0.08 |
ENST00000471583.1
ENST00000487270.1 ENST00000488612.1 |
RAD51B
|
RAD51 paralog B |
| chr1_+_207818460 | 0.08 |
ENST00000508064.2
|
CR1L
|
complement component (3b/4b) receptor 1-like |
| chr1_-_146082633 | 0.08 |
ENST00000605317.1
ENST00000604938.1 ENST00000339388.5 |
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chr1_-_161147275 | 0.08 |
ENST00000319769.5
ENST00000367998.1 |
B4GALT3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
| chr1_-_16939976 | 0.08 |
ENST00000430580.2
|
NBPF1
|
neuroblastoma breakpoint family, member 1 |
| chr2_-_128615681 | 0.08 |
ENST00000409955.1
ENST00000272645.4 |
POLR2D
|
polymerase (RNA) II (DNA directed) polypeptide D |
| chr18_-_51750948 | 0.08 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr7_-_43909090 | 0.07 |
ENST00000317534.5
|
MRPS24
|
mitochondrial ribosomal protein S24 |
| chr12_-_113772835 | 0.07 |
ENST00000552014.1
ENST00000548186.1 ENST00000202831.3 ENST00000549181.1 |
SLC8B1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr3_+_152552685 | 0.07 |
ENST00000305097.3
|
P2RY1
|
purinergic receptor P2Y, G-protein coupled, 1 |
| chr6_-_31697563 | 0.07 |
ENST00000375789.2
ENST00000416410.1 |
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr11_-_72145641 | 0.07 |
ENST00000538039.1
ENST00000445069.2 |
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr1_-_23810664 | 0.07 |
ENST00000336689.3
ENST00000437606.2 |
ASAP3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr1_+_45805342 | 0.07 |
ENST00000372090.5
|
TOE1
|
target of EGR1, member 1 (nuclear) |
| chr9_-_131038266 | 0.07 |
ENST00000490628.1
ENST00000421699.2 ENST00000450617.1 |
GOLGA2
|
golgin A2 |
| chrX_-_103401649 | 0.07 |
ENST00000357421.4
|
SLC25A53
|
solute carrier family 25, member 53 |
| chr2_-_73053126 | 0.07 |
ENST00000272427.6
ENST00000410104.1 |
EXOC6B
|
exocyst complex component 6B |
| chr19_+_44100544 | 0.07 |
ENST00000391965.2
ENST00000525771.1 |
ZNF576
|
zinc finger protein 576 |
| chr9_-_35111570 | 0.07 |
ENST00000378561.1
ENST00000603301.1 |
FAM214B
|
family with sequence similarity 214, member B |
| chr17_+_7184986 | 0.07 |
ENST00000317370.8
ENST00000571308.1 |
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr3_-_138048682 | 0.07 |
ENST00000383180.2
|
NME9
|
NME/NM23 family member 9 |
| chr19_-_56110859 | 0.07 |
ENST00000221665.3
ENST00000592585.1 |
FIZ1
|
FLT3-interacting zinc finger 1 |
| chrX_+_153170455 | 0.06 |
ENST00000430697.1
ENST00000337474.5 ENST00000370049.1 |
AVPR2
|
arginine vasopressin receptor 2 |
| chr14_+_23299088 | 0.06 |
ENST00000355151.5
ENST00000397496.3 ENST00000555345.1 ENST00000432849.3 ENST00000553711.1 ENST00000556465.1 ENST00000397505.2 ENST00000557221.1 ENST00000311892.6 ENST00000556840.1 ENST00000555536.1 |
MRPL52
|
mitochondrial ribosomal protein L52 |
| chr19_+_8509842 | 0.06 |
ENST00000325495.4
ENST00000600092.1 ENST00000594907.1 ENST00000596984.1 ENST00000601645.1 |
HNRNPM
|
heterogeneous nuclear ribonucleoprotein M |
| chr1_+_21766641 | 0.06 |
ENST00000342104.5
|
NBPF3
|
neuroblastoma breakpoint family, member 3 |
| chr7_+_29234028 | 0.06 |
ENST00000222792.6
|
CHN2
|
chimerin 2 |
| chr22_-_41985865 | 0.06 |
ENST00000216259.7
|
PMM1
|
phosphomannomutase 1 |
| chr9_+_72658490 | 0.06 |
ENST00000377182.4
|
MAMDC2
|
MAM domain containing 2 |
| chr19_-_12792020 | 0.06 |
ENST00000594424.1
ENST00000597152.1 ENST00000596162.1 |
DHPS
|
deoxyhypusine synthase |
| chr7_-_98030360 | 0.06 |
ENST00000005260.8
|
BAIAP2L1
|
BAI1-associated protein 2-like 1 |
| chr1_+_162467595 | 0.06 |
ENST00000538489.1
ENST00000489294.1 |
UHMK1
|
U2AF homology motif (UHM) kinase 1 |
| chr12_-_6579808 | 0.06 |
ENST00000535180.1
ENST00000400911.3 |
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr12_-_6580094 | 0.06 |
ENST00000361716.3
|
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr21_+_45905448 | 0.06 |
ENST00000449713.1
|
AP001065.15
|
AP001065.15 |
| chr3_+_132136331 | 0.06 |
ENST00000260818.6
|
DNAJC13
|
DnaJ (Hsp40) homolog, subfamily C, member 13 |
| chr7_-_19748640 | 0.05 |
ENST00000222567.5
|
TWISTNB
|
TWIST neighbor |
| chr19_-_44008863 | 0.05 |
ENST00000601646.1
|
PHLDB3
|
pleckstrin homology-like domain, family B, member 3 |
| chr4_-_4291861 | 0.05 |
ENST00000343470.4
|
LYAR
|
Ly1 antibody reactive |
| chr4_-_4291761 | 0.05 |
ENST00000513174.1
|
LYAR
|
Ly1 antibody reactive |
| chr12_-_124457257 | 0.05 |
ENST00000545891.1
|
CCDC92
|
coiled-coil domain containing 92 |
| chr16_-_30905263 | 0.05 |
ENST00000572628.1
|
BCL7C
|
B-cell CLL/lymphoma 7C |
| chr19_+_36631867 | 0.05 |
ENST00000588780.1
|
CAPNS1
|
calpain, small subunit 1 |
| chr1_+_21766588 | 0.05 |
ENST00000454000.2
ENST00000318220.6 ENST00000318249.5 |
NBPF3
|
neuroblastoma breakpoint family, member 3 |
| chr17_-_6915646 | 0.05 |
ENST00000574377.1
ENST00000399541.2 ENST00000399540.2 ENST00000575727.1 ENST00000573939.1 |
AC027763.2
|
Uncharacterized protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF13
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.3 | GO:0015709 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.1 | 0.4 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.3 | GO:0006050 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.1 | 0.2 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.1 | 1.1 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 0.4 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.2 | GO:0016334 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.1 | 0.3 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.1 | 0.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.2 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.3 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.6 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.4 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.2 | GO:0051344 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.2 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.0 | 0.4 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 | 0.7 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.1 | GO:0002353 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.0 | 0.1 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.3 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.2 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.1 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.0 | 0.1 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.1 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.2 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.7 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 1.1 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.4 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.1 | GO:0035811 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) negative regulation of urine volume(GO:0035811) |
| 0.0 | 0.2 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.2 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.0 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.5 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.7 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.8 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.3 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.4 | GO:0044393 | microspike(GO:0044393) |
| 0.1 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.2 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.7 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.3 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.4 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.1 | 0.6 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.3 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.1 | 0.3 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.1 | 0.3 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.1 | 0.3 | GO:0034038 | deoxyhypusine synthase activity(GO:0034038) |
| 0.1 | 0.7 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.2 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.2 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.1 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.3 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.3 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.0 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.4 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.4 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |