Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
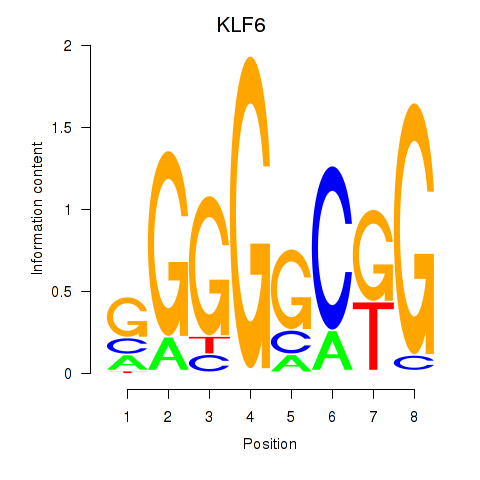
Results for KLF6
Z-value: 0.68
Transcription factors associated with KLF6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF6
|
ENSG00000067082.10 | Kruppel like factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF6 | hg19_v2_chr10_-_3827371_3827386 | -0.10 | 6.3e-01 | Click! |
Activity profile of KLF6 motif
Sorted Z-values of KLF6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_42277203 | 2.21 |
ENST00000587097.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr19_+_45504688 | 1.92 |
ENST00000221452.8
ENST00000540120.1 ENST00000505236.1 |
RELB
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr16_-_11681023 | 1.71 |
ENST00000570904.1
ENST00000574701.1 |
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr1_+_169075554 | 1.58 |
ENST00000367815.4
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr6_-_29595779 | 1.56 |
ENST00000355973.3
ENST00000377012.4 |
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr21_+_43639211 | 1.33 |
ENST00000450121.1
ENST00000398449.3 ENST00000361802.2 |
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr6_+_83073952 | 1.18 |
ENST00000543496.1
|
TPBG
|
trophoblast glycoprotein |
| chr17_-_53499310 | 1.15 |
ENST00000262065.3
|
MMD
|
monocyte to macrophage differentiation-associated |
| chr6_-_143266297 | 1.11 |
ENST00000367603.2
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr9_-_95896550 | 1.06 |
ENST00000375446.4
|
NINJ1
|
ninjurin 1 |
| chr2_+_150187020 | 1.03 |
ENST00000334166.4
|
LYPD6
|
LY6/PLAUR domain containing 6 |
| chr17_-_42276574 | 1.00 |
ENST00000589805.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr6_+_37137939 | 0.99 |
ENST00000373509.5
|
PIM1
|
pim-1 oncogene |
| chr6_-_137113604 | 0.97 |
ENST00000359015.4
|
MAP3K5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr10_-_101380121 | 0.94 |
ENST00000370495.4
|
SLC25A28
|
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr3_+_133292759 | 0.93 |
ENST00000431519.2
|
CDV3
|
CDV3 homolog (mouse) |
| chr11_-_72353451 | 0.92 |
ENST00000376450.3
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated |
| chr6_+_127439749 | 0.90 |
ENST00000356698.4
|
RSPO3
|
R-spondin 3 |
| chr6_+_37787704 | 0.88 |
ENST00000474522.1
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr8_-_101322132 | 0.82 |
ENST00000523481.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr1_-_114696472 | 0.79 |
ENST00000393296.1
ENST00000369547.1 ENST00000610222.1 |
SYT6
|
synaptotagmin VI |
| chr6_+_32811885 | 0.79 |
ENST00000458296.1
ENST00000413039.1 ENST00000429600.1 ENST00000412095.1 ENST00000415067.1 ENST00000395330.1 |
TAPSAR1
PSMB9
|
TAP1 and PSMB8 antisense RNA 1 proteasome (prosome, macropain) subunit, beta type, 9 |
| chr17_+_6926339 | 0.72 |
ENST00000293805.5
|
BCL6B
|
B-cell CLL/lymphoma 6, member B |
| chr17_-_73874654 | 0.72 |
ENST00000254816.2
|
TRIM47
|
tripartite motif containing 47 |
| chr15_-_75744014 | 0.71 |
ENST00000394947.3
ENST00000565264.1 |
SIN3A
|
SIN3 transcription regulator family member A |
| chr15_-_75743915 | 0.70 |
ENST00000394949.4
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr11_-_72492903 | 0.69 |
ENST00000537947.1
|
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr11_+_75526212 | 0.69 |
ENST00000356136.3
|
UVRAG
|
UV radiation resistance associated |
| chr15_-_70388943 | 0.67 |
ENST00000559048.1
ENST00000560939.1 ENST00000440567.3 ENST00000557907.1 ENST00000558379.1 ENST00000451782.2 ENST00000559929.1 |
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr14_+_93897272 | 0.66 |
ENST00000393151.2
|
UNC79
|
unc-79 homolog (C. elegans) |
| chr19_+_13906250 | 0.65 |
ENST00000254323.2
|
ZSWIM4
|
zinc finger, SWIM-type containing 4 |
| chr1_-_205782304 | 0.62 |
ENST00000367137.3
|
SLC41A1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr22_+_31608219 | 0.61 |
ENST00000406516.1
ENST00000444929.2 ENST00000331728.4 |
LIMK2
|
LIM domain kinase 2 |
| chr7_+_116165038 | 0.57 |
ENST00000393470.1
|
CAV1
|
caveolin 1, caveolae protein, 22kDa |
| chr6_+_32811861 | 0.57 |
ENST00000453426.1
|
TAPSAR1
|
TAP1 and PSMB8 antisense RNA 1 |
| chr11_-_3862059 | 0.57 |
ENST00000396978.1
|
RHOG
|
ras homolog family member G |
| chr19_-_47734448 | 0.56 |
ENST00000439096.2
|
BBC3
|
BCL2 binding component 3 |
| chr3_-_88108212 | 0.55 |
ENST00000482016.1
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr2_+_219433588 | 0.54 |
ENST00000295701.5
|
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr4_+_4861385 | 0.53 |
ENST00000382723.4
|
MSX1
|
msh homeobox 1 |
| chr14_+_93897199 | 0.52 |
ENST00000553484.1
|
UNC79
|
unc-79 homolog (C. elegans) |
| chr19_+_34287174 | 0.51 |
ENST00000587559.1
ENST00000588637.1 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr16_+_68678739 | 0.50 |
ENST00000264012.4
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental) |
| chr16_+_68678892 | 0.50 |
ENST00000429102.2
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental) |
| chr5_-_94620239 | 0.49 |
ENST00000515393.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr11_+_64008525 | 0.49 |
ENST00000449942.2
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr2_-_161350305 | 0.48 |
ENST00000348849.3
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1 |
| chr18_-_51750948 | 0.47 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr8_-_101321584 | 0.47 |
ENST00000523167.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr8_-_80680078 | 0.47 |
ENST00000337919.5
ENST00000354724.3 |
HEY1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr4_-_6202247 | 0.46 |
ENST00000409021.3
ENST00000409371.3 |
JAKMIP1
|
janus kinase and microtubule interacting protein 1 |
| chr3_+_133293278 | 0.46 |
ENST00000508481.1
ENST00000420115.2 ENST00000504867.1 ENST00000507408.1 ENST00000511392.1 ENST00000515421.1 |
CDV3
|
CDV3 homolog (mouse) |
| chr11_+_64009072 | 0.46 |
ENST00000535135.1
ENST00000394540.3 |
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr11_+_111473108 | 0.45 |
ENST00000304987.3
|
SIK2
|
salt-inducible kinase 2 |
| chr1_-_111746966 | 0.45 |
ENST00000369752.5
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr15_-_70388599 | 0.43 |
ENST00000560996.1
ENST00000558201.1 |
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr6_+_31588478 | 0.42 |
ENST00000376007.4
ENST00000376033.2 |
PRRC2A
|
proline-rich coiled-coil 2A |
| chr15_+_80987617 | 0.42 |
ENST00000258884.4
ENST00000558464.1 |
ABHD17C
|
abhydrolase domain containing 17C |
| chr11_-_72492878 | 0.42 |
ENST00000535054.1
ENST00000545082.1 |
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr1_-_6321035 | 0.39 |
ENST00000377893.2
|
GPR153
|
G protein-coupled receptor 153 |
| chr10_+_23983671 | 0.39 |
ENST00000376462.1
|
KIAA1217
|
KIAA1217 |
| chr11_-_108464465 | 0.38 |
ENST00000525344.1
|
EXPH5
|
exophilin 5 |
| chr16_+_75033210 | 0.37 |
ENST00000566250.1
ENST00000567962.1 |
ZNRF1
|
zinc and ring finger 1, E3 ubiquitin protein ligase |
| chr19_+_10400615 | 0.37 |
ENST00000221980.4
|
ICAM5
|
intercellular adhesion molecule 5, telencephalin |
| chr2_+_219433281 | 0.36 |
ENST00000273064.6
ENST00000509807.2 ENST00000542068.1 |
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr5_+_67511524 | 0.36 |
ENST00000521381.1
ENST00000521657.1 |
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chrX_+_37430822 | 0.35 |
ENST00000378621.3
ENST00000378619.3 |
LANCL3
|
LanC lantibiotic synthetase component C-like 3 (bacterial) |
| chr9_+_34989638 | 0.35 |
ENST00000453597.3
ENST00000335998.3 ENST00000312316.5 |
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr2_+_111878483 | 0.34 |
ENST00000308659.8
ENST00000357757.2 ENST00000393253.2 ENST00000337565.5 ENST00000393256.3 |
BCL2L11
|
BCL2-like 11 (apoptosis facilitator) |
| chr1_-_156698591 | 0.34 |
ENST00000368219.1
|
ISG20L2
|
interferon stimulated exonuclease gene 20kDa-like 2 |
| chr4_+_8201091 | 0.34 |
ENST00000382521.3
ENST00000245105.3 ENST00000457650.2 ENST00000539824.1 |
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr6_+_44095347 | 0.34 |
ENST00000323267.6
|
TMEM63B
|
transmembrane protein 63B |
| chr9_+_35605274 | 0.34 |
ENST00000336395.5
|
TESK1
|
testis-specific kinase 1 |
| chr16_+_25703274 | 0.34 |
ENST00000331351.5
|
HS3ST4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4 |
| chr6_-_32811771 | 0.34 |
ENST00000395339.3
ENST00000374882.3 |
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr1_-_161102213 | 0.34 |
ENST00000458050.2
|
DEDD
|
death effector domain containing |
| chr20_+_37101455 | 0.34 |
ENST00000262879.6
ENST00000397042.3 ENST00000397038.1 ENST00000537204.1 |
RALGAPB
|
Ral GTPase activating protein, beta subunit (non-catalytic) |
| chr5_+_170846640 | 0.33 |
ENST00000274625.5
|
FGF18
|
fibroblast growth factor 18 |
| chr20_+_37101526 | 0.33 |
ENST00000397040.1
|
RALGAPB
|
Ral GTPase activating protein, beta subunit (non-catalytic) |
| chr12_+_70132632 | 0.32 |
ENST00000378815.6
ENST00000483530.2 ENST00000325555.9 |
RAB3IP
|
RAB3A interacting protein |
| chr19_+_10196981 | 0.32 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr9_+_100174344 | 0.32 |
ENST00000422139.2
|
TDRD7
|
tudor domain containing 7 |
| chr16_+_69599899 | 0.31 |
ENST00000567239.1
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr9_-_37465396 | 0.31 |
ENST00000307750.4
|
ZBTB5
|
zinc finger and BTB domain containing 5 |
| chr17_-_60885700 | 0.31 |
ENST00000583600.1
|
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr20_+_48807351 | 0.31 |
ENST00000303004.3
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr1_-_231004220 | 0.31 |
ENST00000366663.5
|
C1orf198
|
chromosome 1 open reading frame 198 |
| chr22_-_38484922 | 0.30 |
ENST00000428572.1
|
BAIAP2L2
|
BAI1-associated protein 2-like 2 |
| chr12_-_49351148 | 0.30 |
ENST00000398092.4
ENST00000539611.1 |
RP11-302B13.5
ARF3
|
ADP-ribosylation factor 3 ADP-ribosylation factor 3 |
| chr19_+_42724423 | 0.30 |
ENST00000301215.3
ENST00000597945.1 |
ZNF526
|
zinc finger protein 526 |
| chr20_-_31172598 | 0.30 |
ENST00000201961.2
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr11_+_58346584 | 0.30 |
ENST00000316059.6
|
ZFP91
|
ZFP91 zinc finger protein |
| chr21_-_35987438 | 0.29 |
ENST00000313806.4
|
RCAN1
|
regulator of calcineurin 1 |
| chr17_-_60885659 | 0.29 |
ENST00000311269.5
|
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr3_+_38495333 | 0.29 |
ENST00000352511.4
|
ACVR2B
|
activin A receptor, type IIB |
| chr10_+_35415851 | 0.29 |
ENST00000374726.3
|
CREM
|
cAMP responsive element modulator |
| chr11_+_66886717 | 0.29 |
ENST00000398645.2
|
KDM2A
|
lysine (K)-specific demethylase 2A |
| chr1_+_107683436 | 0.28 |
ENST00000370068.1
|
NTNG1
|
netrin G1 |
| chr6_+_99282570 | 0.28 |
ENST00000328345.5
|
POU3F2
|
POU class 3 homeobox 2 |
| chr18_-_48723690 | 0.28 |
ENST00000406189.3
|
MEX3C
|
mex-3 RNA binding family member C |
| chr17_-_7120498 | 0.28 |
ENST00000485100.1
|
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr19_+_14672755 | 0.28 |
ENST00000594545.1
|
TECR
|
trans-2,3-enoyl-CoA reductase |
| chr15_+_52311398 | 0.28 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr6_-_31926629 | 0.28 |
ENST00000375425.5
ENST00000426722.1 ENST00000441998.1 ENST00000444811.2 ENST00000375429.3 |
NELFE
|
negative elongation factor complex member E |
| chr12_+_57482665 | 0.27 |
ENST00000300131.3
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr2_-_25475120 | 0.27 |
ENST00000380746.4
ENST00000402667.1 |
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha |
| chr5_+_38845960 | 0.27 |
ENST00000502536.1
|
OSMR
|
oncostatin M receptor |
| chr18_-_12884150 | 0.27 |
ENST00000591115.1
ENST00000309660.5 |
PTPN2
|
protein tyrosine phosphatase, non-receptor type 2 |
| chr13_-_27745936 | 0.27 |
ENST00000282344.6
|
USP12
|
ubiquitin specific peptidase 12 |
| chr10_-_75255668 | 0.27 |
ENST00000545874.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr20_+_30555805 | 0.26 |
ENST00000562532.2
|
XKR7
|
XK, Kell blood group complex subunit-related family, member 7 |
| chr7_+_116593292 | 0.26 |
ENST00000393446.2
ENST00000265437.5 ENST00000393451.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr4_-_174451370 | 0.26 |
ENST00000359562.4
|
HAND2
|
heart and neural crest derivatives expressed 2 |
| chr17_-_28257080 | 0.26 |
ENST00000579954.1
ENST00000540801.1 ENST00000269033.3 ENST00000590153.1 ENST00000582084.1 |
SSH2
|
slingshot protein phosphatase 2 |
| chr6_+_36164487 | 0.25 |
ENST00000357641.6
|
BRPF3
|
bromodomain and PHD finger containing, 3 |
| chr2_-_161349909 | 0.25 |
ENST00000392753.3
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1 |
| chr16_+_75032901 | 0.25 |
ENST00000335325.4
ENST00000320619.6 |
ZNRF1
|
zinc and ring finger 1, E3 ubiquitin protein ligase |
| chr2_+_27071045 | 0.25 |
ENST00000401478.1
|
DPYSL5
|
dihydropyrimidinase-like 5 |
| chr12_-_120554622 | 0.25 |
ENST00000229340.5
|
RAB35
|
RAB35, member RAS oncogene family |
| chr1_+_114472481 | 0.25 |
ENST00000369555.2
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr6_+_31126291 | 0.25 |
ENST00000376257.3
ENST00000376255.4 |
TCF19
|
transcription factor 19 |
| chr22_-_17602143 | 0.25 |
ENST00000331437.3
|
CECR6
|
cat eye syndrome chromosome region, candidate 6 |
| chr6_-_35656712 | 0.24 |
ENST00000357266.4
ENST00000542713.1 |
FKBP5
|
FK506 binding protein 5 |
| chr16_+_68679193 | 0.24 |
ENST00000581171.1
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental) |
| chr18_-_12884259 | 0.24 |
ENST00000353319.4
ENST00000327283.3 |
PTPN2
|
protein tyrosine phosphatase, non-receptor type 2 |
| chr3_+_10290596 | 0.24 |
ENST00000448281.2
|
TATDN2
|
TatD DNase domain containing 2 |
| chr19_-_41196534 | 0.24 |
ENST00000252891.4
|
NUMBL
|
numb homolog (Drosophila)-like |
| chr2_+_220299547 | 0.24 |
ENST00000312358.7
|
SPEG
|
SPEG complex locus |
| chr6_-_82462425 | 0.24 |
ENST00000369754.3
ENST00000320172.6 ENST00000369756.3 |
FAM46A
|
family with sequence similarity 46, member A |
| chr6_-_35656685 | 0.24 |
ENST00000539068.1
ENST00000540787.1 |
FKBP5
|
FK506 binding protein 5 |
| chr7_-_100493482 | 0.24 |
ENST00000411582.1
ENST00000419336.2 ENST00000241069.5 ENST00000302913.4 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr2_-_114514181 | 0.23 |
ENST00000409342.1
|
SLC35F5
|
solute carrier family 35, member F5 |
| chr15_-_100273544 | 0.23 |
ENST00000409796.1
ENST00000545021.1 ENST00000344791.2 ENST00000332728.4 ENST00000450512.1 |
LYSMD4
|
LysM, putative peptidoglycan-binding, domain containing 4 |
| chr19_-_10764509 | 0.23 |
ENST00000591501.1
|
ILF3-AS1
|
ILF3 antisense RNA 1 (head to head) |
| chr1_+_107682629 | 0.23 |
ENST00000370074.4
ENST00000370073.2 ENST00000370071.2 ENST00000542803.1 ENST00000370061.3 ENST00000370072.3 ENST00000370070.2 |
NTNG1
|
netrin G1 |
| chr3_+_10289707 | 0.23 |
ENST00000287652.4
|
TATDN2
|
TatD DNase domain containing 2 |
| chr19_-_41220957 | 0.23 |
ENST00000596357.1
ENST00000243583.6 ENST00000600080.1 ENST00000595254.1 ENST00000601967.1 |
ADCK4
|
aarF domain containing kinase 4 |
| chr12_-_120554534 | 0.23 |
ENST00000538903.1
ENST00000534951.1 |
RAB35
|
RAB35, member RAS oncogene family |
| chr9_-_131038266 | 0.23 |
ENST00000490628.1
ENST00000421699.2 ENST00000450617.1 |
GOLGA2
|
golgin A2 |
| chr19_-_10444188 | 0.23 |
ENST00000293677.6
|
RAVER1
|
ribonucleoprotein, PTB-binding 1 |
| chr1_-_156698181 | 0.22 |
ENST00000313146.6
|
ISG20L2
|
interferon stimulated exonuclease gene 20kDa-like 2 |
| chr11_-_3862206 | 0.22 |
ENST00000351018.4
|
RHOG
|
ras homolog family member G |
| chr17_+_36861735 | 0.22 |
ENST00000378137.5
ENST00000325718.7 |
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr5_-_142782862 | 0.22 |
ENST00000415690.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr7_+_120628731 | 0.22 |
ENST00000310396.5
|
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr17_-_7518145 | 0.21 |
ENST00000250113.7
ENST00000571597.1 |
FXR2
|
fragile X mental retardation, autosomal homolog 2 |
| chrX_-_49056635 | 0.21 |
ENST00000472598.1
ENST00000538567.1 ENST00000479808.1 ENST00000263233.4 |
SYP
|
synaptophysin |
| chr1_-_226924980 | 0.21 |
ENST00000272117.3
|
ITPKB
|
inositol-trisphosphate 3-kinase B |
| chr12_-_49351228 | 0.21 |
ENST00000541959.1
ENST00000447318.2 |
ARF3
|
ADP-ribosylation factor 3 |
| chr6_+_111195973 | 0.21 |
ENST00000368885.3
ENST00000368882.3 ENST00000451850.2 ENST00000368877.5 |
AMD1
|
adenosylmethionine decarboxylase 1 |
| chr22_-_17602200 | 0.20 |
ENST00000399875.1
|
CECR6
|
cat eye syndrome chromosome region, candidate 6 |
| chr17_-_74449252 | 0.20 |
ENST00000319380.7
|
UBE2O
|
ubiquitin-conjugating enzyme E2O |
| chr5_+_56111361 | 0.20 |
ENST00000399503.3
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr6_+_1389989 | 0.20 |
ENST00000259806.1
|
FOXF2
|
forkhead box F2 |
| chr7_+_143078652 | 0.20 |
ENST00000354434.4
ENST00000449423.2 |
ZYX
|
zyxin |
| chr2_-_27603582 | 0.20 |
ENST00000323703.6
ENST00000436006.1 |
ZNF513
|
zinc finger protein 513 |
| chr17_+_28256874 | 0.20 |
ENST00000541045.1
ENST00000536908.2 |
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr14_-_73925225 | 0.20 |
ENST00000356296.4
ENST00000355058.3 ENST00000359560.3 ENST00000557597.1 ENST00000554394.1 ENST00000555238.1 ENST00000535282.1 ENST00000555987.1 ENST00000555394.1 ENST00000554546.1 |
NUMB
|
numb homolog (Drosophila) |
| chr14_-_39572345 | 0.19 |
ENST00000548032.2
ENST00000556092.1 ENST00000557280.1 ENST00000545328.2 ENST00000553970.1 |
SEC23A
|
Sec23 homolog A (S. cerevisiae) |
| chr17_+_7452442 | 0.19 |
ENST00000557233.1
|
TNFSF12
|
tumor necrosis factor (ligand) superfamily, member 12 |
| chr15_-_66084428 | 0.19 |
ENST00000443035.3
ENST00000431932.2 |
DENND4A
|
DENN/MADD domain containing 4A |
| chr17_+_7452336 | 0.19 |
ENST00000293826.4
|
TNFSF12-TNFSF13
|
TNFSF12-TNFSF13 readthrough |
| chr5_-_168006591 | 0.19 |
ENST00000239231.6
|
PANK3
|
pantothenate kinase 3 |
| chr17_-_74733404 | 0.19 |
ENST00000508921.3
ENST00000583836.1 ENST00000358156.6 ENST00000392485.2 ENST00000359995.5 |
SRSF2
|
serine/arginine-rich splicing factor 2 |
| chr4_+_184426147 | 0.19 |
ENST00000302327.3
|
ING2
|
inhibitor of growth family, member 2 |
| chr8_-_125486755 | 0.19 |
ENST00000499418.2
ENST00000530778.1 |
RNF139-AS1
|
RNF139 antisense RNA 1 (head to head) |
| chr2_-_30144432 | 0.18 |
ENST00000389048.3
|
ALK
|
anaplastic lymphoma receptor tyrosine kinase |
| chr19_-_41196458 | 0.18 |
ENST00000598779.1
|
NUMBL
|
numb homolog (Drosophila)-like |
| chr20_+_57466629 | 0.18 |
ENST00000371081.1
ENST00000338783.6 |
GNAS
|
GNAS complex locus |
| chr1_-_23670817 | 0.18 |
ENST00000478691.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr6_-_31926208 | 0.18 |
ENST00000454913.1
ENST00000436289.2 |
NELFE
|
negative elongation factor complex member E |
| chr1_+_6845384 | 0.18 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr12_+_57482877 | 0.18 |
ENST00000342556.6
ENST00000357680.4 |
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr12_-_31479107 | 0.18 |
ENST00000542983.1
|
FAM60A
|
family with sequence similarity 60, member A |
| chr9_-_101984184 | 0.18 |
ENST00000476832.1
|
ALG2
|
ALG2, alpha-1,3/1,6-mannosyltransferase |
| chr1_-_38273840 | 0.18 |
ENST00000373044.2
|
YRDC
|
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr22_+_45680822 | 0.18 |
ENST00000216211.4
ENST00000396082.2 |
UPK3A
|
uroplakin 3A |
| chr17_-_1389228 | 0.18 |
ENST00000438665.2
|
MYO1C
|
myosin IC |
| chr7_+_143079000 | 0.18 |
ENST00000392910.2
|
ZYX
|
zyxin |
| chr11_+_45907177 | 0.17 |
ENST00000241014.2
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr10_+_124768482 | 0.17 |
ENST00000368869.4
ENST00000358776.4 |
ACADSB
|
acyl-CoA dehydrogenase, short/branched chain |
| chr4_+_183370146 | 0.17 |
ENST00000510504.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr16_+_50582222 | 0.17 |
ENST00000268459.3
|
NKD1
|
naked cuticle homolog 1 (Drosophila) |
| chr1_-_38325256 | 0.17 |
ENST00000373036.4
|
MTF1
|
metal-regulatory transcription factor 1 |
| chr16_+_67465016 | 0.17 |
ENST00000326152.5
|
HSD11B2
|
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr12_+_124196865 | 0.17 |
ENST00000330342.3
|
ATP6V0A2
|
ATPase, H+ transporting, lysosomal V0 subunit a2 |
| chr20_+_35201857 | 0.17 |
ENST00000373874.2
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr3_-_113415441 | 0.17 |
ENST00000491165.1
ENST00000316407.4 |
KIAA2018
|
KIAA2018 |
| chr17_-_42143963 | 0.17 |
ENST00000585388.1
ENST00000293406.3 |
LSM12
|
LSM12 homolog (S. cerevisiae) |
| chr19_+_10828724 | 0.16 |
ENST00000585892.1
ENST00000314646.5 ENST00000359692.6 |
DNM2
|
dynamin 2 |
| chr5_-_142783175 | 0.16 |
ENST00000231509.3
ENST00000394464.2 |
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr7_-_130353553 | 0.16 |
ENST00000330992.7
ENST00000445977.2 |
COPG2
|
coatomer protein complex, subunit gamma 2 |
| chr9_-_120177342 | 0.16 |
ENST00000361209.2
|
ASTN2
|
astrotactin 2 |
| chr14_-_81687575 | 0.16 |
ENST00000434192.2
|
GTF2A1
|
general transcription factor IIA, 1, 19/37kDa |
| chrX_+_56590002 | 0.16 |
ENST00000338222.5
|
UBQLN2
|
ubiquilin 2 |
| chr1_-_38512450 | 0.16 |
ENST00000373012.2
|
POU3F1
|
POU class 3 homeobox 1 |
| chr11_+_45868957 | 0.16 |
ENST00000443527.2
|
CRY2
|
cryptochrome 2 (photolyase-like) |
| chr2_+_105471969 | 0.16 |
ENST00000361360.2
|
POU3F3
|
POU class 3 homeobox 3 |
| chr11_+_65479462 | 0.16 |
ENST00000377046.3
ENST00000352980.4 ENST00000341318.4 |
KAT5
|
K(lysine) acetyltransferase 5 |
| chr14_+_23790690 | 0.16 |
ENST00000556821.1
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0032773 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.4 | 1.5 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.3 | 1.3 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.3 | 1.6 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 0.3 | 0.9 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.2 | 0.9 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.2 | 0.6 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.2 | 0.6 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.2 | 0.5 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.2 | 0.5 | GO:1902214 | regulation of interleukin-4-mediated signaling pathway(GO:1902214) negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.1 | 0.8 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.8 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 0.4 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 0.5 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.1 | 0.3 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 1.8 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 1.1 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.1 | 0.7 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.1 | 0.9 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.1 | 2.4 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.2 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 | 1.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.5 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 1.2 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 0.4 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.1 | 1.1 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.1 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.3 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.3 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.1 | 0.2 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.1 | 1.9 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 | 1.0 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.1 | 0.2 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 0.2 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.4 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.4 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.3 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.4 | GO:0090306 | Golgi disassembly(GO:0090166) spindle assembly involved in meiosis(GO:0090306) |
| 0.0 | 0.1 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.0 | 0.3 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.3 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.0 | 0.1 | GO:1903572 | coronary vein morphogenesis(GO:0003169) cardiac vascular smooth muscle cell development(GO:0060948) regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.2 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.0 | 1.6 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.3 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.0 | 0.2 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.2 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.1 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.0 | 0.1 | GO:0072086 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.2 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.2 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.3 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.1 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.4 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.0 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.2 | GO:2000828 | post-embryonic body morphogenesis(GO:0040032) regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.3 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.2 | GO:0015840 | urea transport(GO:0015840) urinary bladder development(GO:0060157) |
| 0.0 | 0.3 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.8 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 1.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.5 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.4 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 1.2 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.1 | GO:0032240 | regulation of chromatin assembly(GO:0010847) mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) positive regulation of chromatin assembly or disassembly(GO:0045799) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.0 | 1.4 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.0 | 0.2 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.0 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 1.1 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.4 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.1 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.8 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.1 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.0 | 0.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.1 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.0 | 0.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.3 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.1 | GO:0072104 | glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.6 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.2 | GO:0048339 | paraxial mesoderm development(GO:0048339) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.0 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.0 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 0.0 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.2 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.2 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 1.1 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 1.3 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 0.0 | GO:0021747 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) cochlear nucleus development(GO:0021747) negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.1 | GO:0072178 | nephric duct morphogenesis(GO:0072178) |
| 0.0 | 0.5 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.1 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.1 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.1 | GO:0075713 | establishment of integrated proviral latency(GO:0075713) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.2 | 1.8 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.2 | 3.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.2 | 1.0 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.2 | 1.9 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 1.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 1.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.5 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.3 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.1 | 0.4 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 1.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.3 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 1.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.3 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.6 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.7 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.8 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.4 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0032279 | axonemal microtubule(GO:0005879) asymmetric synapse(GO:0032279) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.0 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.3 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.0 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.3 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 0.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.4 | 1.6 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.3 | 1.0 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 0.6 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.2 | 0.9 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.5 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.1 | 0.5 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 0.3 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.3 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 1.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.4 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 0.4 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 0.3 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.1 | 0.4 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.1 | 0.2 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 1.0 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.3 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.9 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 1.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.4 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 3.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.2 | GO:0002060 | purine nucleobase binding(GO:0002060) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0031177 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.0 | 1.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0070362 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.0 | 0.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 0.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 1.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.5 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.3 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 1.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 1.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0052724 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) inositol hexakisphosphate kinase activity(GO:0000828) inositol heptakisphosphate kinase activity(GO:0000829) inositol hexakisphosphate 5-kinase activity(GO:0000832) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 1.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 1.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.1 | GO:0016416 | O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0070573 | peptidyl-dipeptidase activity(GO:0008241) bradykinin receptor binding(GO:0031711) metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.9 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 2.2 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.0 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.6 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 1.7 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.3 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.6 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.6 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.5 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.1 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |