Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for KLF7
Z-value: 0.30
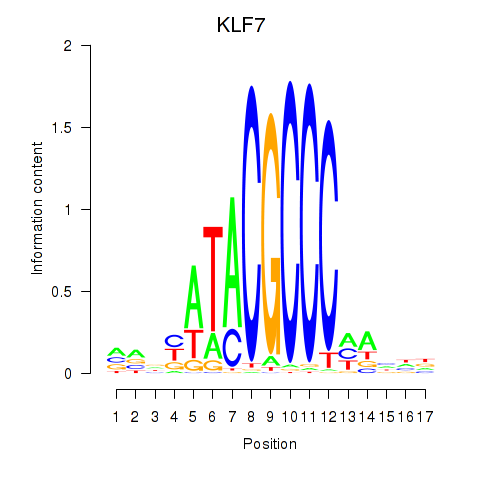
Transcription factors associated with KLF7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF7
|
ENSG00000118263.10 | Kruppel like factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF7 | hg19_v2_chr2_-_208031943_208031991 | 0.22 | 2.8e-01 | Click! |
Activity profile of KLF7 motif
Sorted Z-values of KLF7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_161102367 | 0.95 |
ENST00000464113.1
|
DEDD
|
death effector domain containing |
| chr1_-_161102421 | 0.75 |
ENST00000490843.2
ENST00000368006.3 ENST00000392188.1 ENST00000545495.1 |
DEDD
|
death effector domain containing |
| chr17_-_18218237 | 0.52 |
ENST00000542570.1
|
TOP3A
|
topoisomerase (DNA) III alpha |
| chr15_+_92937144 | 0.34 |
ENST00000539113.1
ENST00000555434.1 |
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr3_+_105085734 | 0.33 |
ENST00000306107.5
|
ALCAM
|
activated leukocyte cell adhesion molecule |
| chr8_+_17104539 | 0.30 |
ENST00000521829.1
ENST00000521005.1 |
VPS37A
|
vacuolar protein sorting 37 homolog A (S. cerevisiae) |
| chr15_+_92937058 | 0.30 |
ENST00000268164.3
|
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr6_+_43603552 | 0.29 |
ENST00000372171.4
|
MAD2L1BP
|
MAD2L1 binding protein |
| chr17_+_18218587 | 0.26 |
ENST00000406438.3
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8 |
| chr1_-_157108266 | 0.24 |
ENST00000326786.4
|
ETV3
|
ets variant 3 |
| chr19_+_39390587 | 0.23 |
ENST00000572515.1
ENST00000392079.3 ENST00000575359.1 ENST00000313582.5 |
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr19_+_39390320 | 0.22 |
ENST00000576510.1
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr17_-_19281203 | 0.19 |
ENST00000487415.2
|
B9D1
|
B9 protein domain 1 |
| chr17_-_18218270 | 0.19 |
ENST00000321105.5
|
TOP3A
|
topoisomerase (DNA) III alpha |
| chr5_+_173315283 | 0.19 |
ENST00000265085.5
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr1_-_21948906 | 0.17 |
ENST00000374761.2
ENST00000599760.1 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr13_-_108867101 | 0.16 |
ENST00000356922.4
|
LIG4
|
ligase IV, DNA, ATP-dependent |
| chr9_+_130565487 | 0.15 |
ENST00000373225.3
ENST00000431857.1 |
FPGS
|
folylpolyglutamate synthase |
| chr6_-_109703600 | 0.14 |
ENST00000512821.1
|
CD164
|
CD164 molecule, sialomucin |
| chr9_+_130565147 | 0.13 |
ENST00000373247.2
ENST00000373245.1 ENST00000393706.2 ENST00000373228.1 |
FPGS
|
folylpolyglutamate synthase |
| chr12_-_49582593 | 0.12 |
ENST00000295766.5
|
TUBA1A
|
tubulin, alpha 1a |
| chr2_-_74710078 | 0.11 |
ENST00000290418.4
|
CCDC142
|
coiled-coil domain containing 142 |
| chr14_+_29234870 | 0.11 |
ENST00000382535.3
|
FOXG1
|
forkhead box G1 |
| chr6_-_109703634 | 0.11 |
ENST00000324953.5
ENST00000310786.4 ENST00000275080.7 ENST00000413644.2 |
CD164
|
CD164 molecule, sialomucin |
| chr8_+_72587535 | 0.10 |
ENST00000519840.1
ENST00000521131.1 |
RP11-1144P22.1
|
RP11-1144P22.1 |
| chr5_+_141348721 | 0.10 |
ENST00000507163.1
ENST00000394519.1 |
RNF14
|
ring finger protein 14 |
| chr6_-_109703663 | 0.10 |
ENST00000368961.5
|
CD164
|
CD164 molecule, sialomucin |
| chr16_+_31539197 | 0.10 |
ENST00000564707.1
|
AHSP
|
alpha hemoglobin stabilizing protein |
| chr3_+_178866199 | 0.10 |
ENST00000263967.3
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr6_-_33239712 | 0.07 |
ENST00000436044.2
|
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr6_+_31865552 | 0.07 |
ENST00000469372.1
ENST00000497706.1 |
C2
|
complement component 2 |
| chr19_-_7694417 | 0.06 |
ENST00000358368.4
ENST00000534844.1 |
XAB2
|
XPA binding protein 2 |
| chr17_-_4269768 | 0.05 |
ENST00000396981.2
|
UBE2G1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr8_-_17104356 | 0.05 |
ENST00000361272.4
ENST00000523917.1 |
CNOT7
|
CCR4-NOT transcription complex, subunit 7 |
| chr17_+_38375574 | 0.03 |
ENST00000323571.4
ENST00000585043.1 ENST00000394103.3 ENST00000536600.1 |
WIPF2
|
WAS/WASL interacting protein family, member 2 |
| chr16_+_81772633 | 0.03 |
ENST00000566191.1
ENST00000565272.1 ENST00000563954.1 ENST00000565054.1 |
RP11-960L18.1
PLCG2
|
RP11-960L18.1 phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| chr5_+_141348598 | 0.03 |
ENST00000394520.2
ENST00000347642.3 |
RNF14
|
ring finger protein 14 |
| chr2_-_188430478 | 0.03 |
ENST00000421427.1
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr22_-_30987849 | 0.02 |
ENST00000402284.3
ENST00000354694.7 |
PES1
|
pescadillo ribosomal biogenesis factor 1 |
| chrX_-_92928557 | 0.02 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr3_+_186353756 | 0.01 |
ENST00000431018.1
ENST00000450521.1 ENST00000539949.1 |
FETUB
|
fetuin B |
| chr2_+_97779233 | 0.01 |
ENST00000461153.2
ENST00000420699.2 |
ANKRD36
|
ankyrin repeat domain 36 |
| chr22_-_30987837 | 0.01 |
ENST00000335214.6
|
PES1
|
pescadillo ribosomal biogenesis factor 1 |
| chr6_-_33239612 | 0.01 |
ENST00000482399.1
ENST00000445902.2 |
VPS52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr19_-_39390350 | 0.01 |
ENST00000447739.1
ENST00000358931.5 ENST00000407552.1 |
SIRT2
|
sirtuin 2 |
| chr1_-_6453399 | 0.00 |
ENST00000608083.1
|
ACOT7
|
acyl-CoA thioesterase 7 |
| chr16_+_31539183 | 0.00 |
ENST00000302312.4
|
AHSP
|
alpha hemoglobin stabilizing protein |
| chr6_+_33239787 | 0.00 |
ENST00000439602.2
ENST00000474973.1 |
RPS18
|
ribosomal protein S18 |
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 0.2 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.1 | 0.2 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.0 | 0.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.7 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.3 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.1 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.0 | 0.2 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 1.7 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.2 | GO:0032807 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.0 | GO:0042025 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.7 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.2 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.2 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.7 | PID ATM PATHWAY | ATM pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.4 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |