Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for LMX1B_MNX1_RAX2
Z-value: 0.47
Transcription factors associated with LMX1B_MNX1_RAX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
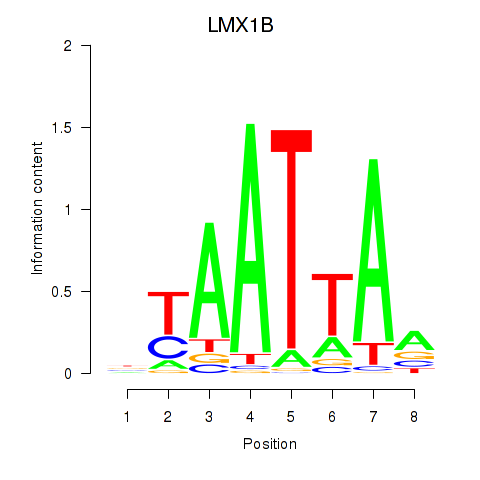
LMX1B
|
ENSG00000136944.13 | LIM homeobox transcription factor 1 beta |
|
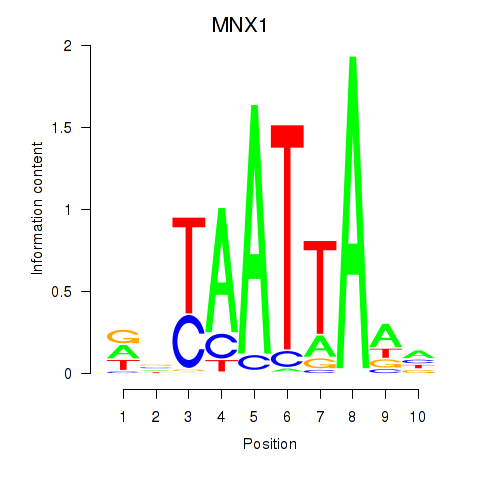
MNX1
|
ENSG00000130675.10 | motor neuron and pancreas homeobox 1 |
|
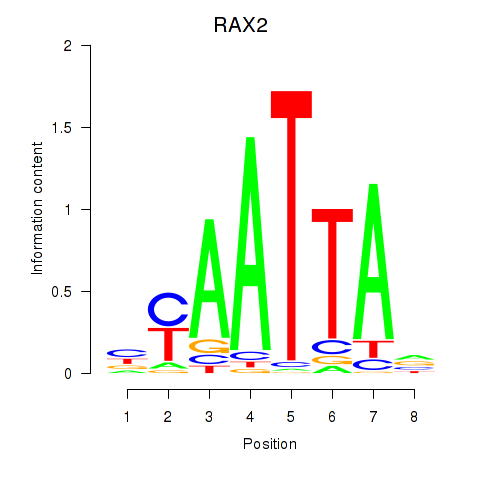
RAX2
|
ENSG00000173976.11 | retina and anterior neural fold homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LMX1B | hg19_v2_chr9_+_129376722_129376748 | -0.19 | 3.6e-01 | Click! |
| MNX1 | hg19_v2_chr7_-_156803329_156803362 | -0.07 | 7.4e-01 | Click! |
| RAX2 | hg19_v2_chr19_-_3772209_3772236 | -0.02 | 9.2e-01 | Click! |
Activity profile of LMX1B_MNX1_RAX2 motif
Sorted Z-values of LMX1B_MNX1_RAX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_76944621 | 1.34 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr7_-_92777606 | 1.21 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr12_+_26348582 | 0.95 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr2_+_152214098 | 0.65 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr1_-_150738261 | 0.64 |
ENST00000448301.2
ENST00000368985.3 |
CTSS
|
cathepsin S |
| chr3_+_173116225 | 0.62 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr2_-_136678123 | 0.58 |
ENST00000422708.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr17_+_47448102 | 0.56 |
ENST00000576461.1
|
RP11-81K2.1
|
Uncharacterized protein |
| chr4_+_169418195 | 0.54 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr6_-_87804815 | 0.54 |
ENST00000369582.2
|
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr11_-_124190184 | 0.54 |
ENST00000357438.2
|
OR8D2
|
olfactory receptor, family 8, subfamily D, member 2 |
| chr5_-_24645078 | 0.54 |
ENST00000264463.4
|
CDH10
|
cadherin 10, type 2 (T2-cadherin) |
| chr14_-_51027838 | 0.53 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr3_-_191000172 | 0.52 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr12_-_28122980 | 0.48 |
ENST00000395868.3
ENST00000534890.1 |
PTHLH
|
parathyroid hormone-like hormone |
| chr4_+_169013666 | 0.47 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr3_-_157221380 | 0.46 |
ENST00000468233.1
|
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr7_-_27169801 | 0.45 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr1_-_101360331 | 0.41 |
ENST00000416479.1
ENST00000370113.3 |
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr3_-_79816965 | 0.41 |
ENST00000464233.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr7_-_22234381 | 0.40 |
ENST00000458533.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chrX_-_21676442 | 0.37 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr11_+_33061543 | 0.37 |
ENST00000432887.1
ENST00000528898.1 ENST00000531632.2 |
TCP11L1
|
t-complex 11, testis-specific-like 1 |
| chr3_-_157221357 | 0.36 |
ENST00000494677.1
|
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr1_-_152131703 | 0.35 |
ENST00000316073.3
|
RPTN
|
repetin |
| chr18_+_32173276 | 0.35 |
ENST00000591816.1
ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA
|
dystrobrevin, alpha |
| chr7_+_134464376 | 0.34 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr12_+_41831485 | 0.34 |
ENST00000539469.2
ENST00000298919.7 |
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr7_-_92855762 | 0.34 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr1_-_237167718 | 0.33 |
ENST00000464121.2
|
MT1HL1
|
metallothionein 1H-like 1 |
| chr1_-_168464875 | 0.33 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr13_-_46716969 | 0.33 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr17_+_41363854 | 0.32 |
ENST00000588693.1
ENST00000588659.1 ENST00000541594.1 ENST00000536052.1 ENST00000331615.3 |
TMEM106A
|
transmembrane protein 106A |
| chr16_+_53133070 | 0.32 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr4_-_39979576 | 0.32 |
ENST00000303538.8
ENST00000503396.1 |
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr3_+_130569429 | 0.31 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr8_+_105235572 | 0.28 |
ENST00000523362.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr2_-_145277569 | 0.28 |
ENST00000303660.4
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr3_+_111717511 | 0.27 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr8_-_109799793 | 0.27 |
ENST00000297459.3
|
TMEM74
|
transmembrane protein 74 |
| chr1_+_160370344 | 0.27 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr3_+_111717600 | 0.27 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr18_-_71959159 | 0.26 |
ENST00000494131.2
ENST00000397914.4 ENST00000340533.4 |
CYB5A
|
cytochrome b5 type A (microsomal) |
| chr2_+_103035102 | 0.26 |
ENST00000264260.2
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr6_+_105404899 | 0.26 |
ENST00000345080.4
|
LIN28B
|
lin-28 homolog B (C. elegans) |
| chr3_+_111718036 | 0.26 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr8_+_107460147 | 0.26 |
ENST00000442977.2
|
OXR1
|
oxidation resistance 1 |
| chr1_+_107683436 | 0.26 |
ENST00000370068.1
|
NTNG1
|
netrin G1 |
| chr17_-_64225508 | 0.25 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr8_+_50824233 | 0.25 |
ENST00000522124.1
|
SNTG1
|
syntrophin, gamma 1 |
| chr12_-_28123206 | 0.25 |
ENST00000542963.1
ENST00000535992.1 |
PTHLH
|
parathyroid hormone-like hormone |
| chr19_-_56110859 | 0.24 |
ENST00000221665.3
ENST00000592585.1 |
FIZ1
|
FLT3-interacting zinc finger 1 |
| chr18_-_33709268 | 0.23 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr4_-_139163491 | 0.23 |
ENST00000280612.5
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chrX_-_110655306 | 0.23 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr3_-_151034734 | 0.22 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chr5_+_31193847 | 0.22 |
ENST00000514738.1
ENST00000265071.2 |
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney) |
| chr1_+_68150744 | 0.22 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr15_+_93443419 | 0.21 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr11_-_102576537 | 0.21 |
ENST00000260229.4
|
MMP27
|
matrix metallopeptidase 27 |
| chr2_-_61697862 | 0.21 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr5_+_125758865 | 0.21 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr5_+_125758813 | 0.21 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr21_-_42219065 | 0.20 |
ENST00000400454.1
|
DSCAM
|
Down syndrome cell adhesion molecule |
| chr6_-_161085291 | 0.20 |
ENST00000316300.5
|
LPA
|
lipoprotein, Lp(a) |
| chr2_-_99871570 | 0.20 |
ENST00000333017.2
ENST00000409679.1 ENST00000423306.1 |
LYG2
|
lysozyme G-like 2 |
| chr10_+_118083919 | 0.20 |
ENST00000333254.3
|
CCDC172
|
coiled-coil domain containing 172 |
| chr1_+_101003687 | 0.19 |
ENST00000315033.4
|
GPR88
|
G protein-coupled receptor 88 |
| chr4_+_114214125 | 0.19 |
ENST00000509550.1
|
ANK2
|
ankyrin 2, neuronal |
| chr17_-_39623681 | 0.18 |
ENST00000225899.3
|
KRT32
|
keratin 32 |
| chr15_-_55562582 | 0.18 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr3_-_33686925 | 0.18 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr10_-_24770632 | 0.18 |
ENST00000596413.1
|
AL353583.1
|
AL353583.1 |
| chr12_+_26348246 | 0.18 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr7_+_134464414 | 0.18 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr1_+_155023757 | 0.18 |
ENST00000356955.2
ENST00000449910.2 ENST00000359280.4 ENST00000360674.4 ENST00000368412.3 ENST00000355956.2 ENST00000368410.2 ENST00000271836.6 ENST00000368413.1 ENST00000531455.1 ENST00000447332.3 |
ADAM15
|
ADAM metallopeptidase domain 15 |
| chr12_-_86650045 | 0.18 |
ENST00000604798.1
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr3_-_108248169 | 0.17 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr15_-_55562479 | 0.17 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr15_-_55563072 | 0.17 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr8_+_24298531 | 0.17 |
ENST00000175238.6
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr8_+_24298597 | 0.17 |
ENST00000380789.1
|
ADAM7
|
ADAM metallopeptidase domain 7 |
| chr2_-_208031943 | 0.16 |
ENST00000421199.1
ENST00000457962.1 |
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr21_+_35736302 | 0.16 |
ENST00000290310.3
|
KCNE2
|
potassium voltage-gated channel, Isk-related family, member 2 |
| chr2_-_145278475 | 0.15 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr5_+_66300446 | 0.15 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr3_-_157824292 | 0.15 |
ENST00000483851.2
|
SHOX2
|
short stature homeobox 2 |
| chr12_+_81110684 | 0.15 |
ENST00000228644.3
|
MYF5
|
myogenic factor 5 |
| chr12_+_93096619 | 0.15 |
ENST00000397833.3
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr3_-_141747950 | 0.15 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr4_+_71458012 | 0.15 |
ENST00000449493.2
|
AMBN
|
ameloblastin (enamel matrix protein) |
| chr1_+_50569575 | 0.14 |
ENST00000371827.1
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr9_+_82188077 | 0.14 |
ENST00000425506.1
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr1_-_190446759 | 0.14 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr2_+_190541153 | 0.13 |
ENST00000313581.4
ENST00000438402.2 ENST00000431575.2 ENST00000281412.6 |
ANKAR
|
ankyrin and armadillo repeat containing |
| chr5_-_147286065 | 0.13 |
ENST00000318315.4
ENST00000515291.1 |
C5orf46
|
chromosome 5 open reading frame 46 |
| chr1_+_107683644 | 0.13 |
ENST00000370067.1
|
NTNG1
|
netrin G1 |
| chr4_+_71457970 | 0.13 |
ENST00000322937.6
|
AMBN
|
ameloblastin (enamel matrix protein) |
| chr15_+_75080883 | 0.13 |
ENST00000567571.1
|
CSK
|
c-src tyrosine kinase |
| chr8_+_38831683 | 0.13 |
ENST00000302495.4
|
HTRA4
|
HtrA serine peptidase 4 |
| chr10_+_24497704 | 0.13 |
ENST00000376456.4
ENST00000458595.1 |
KIAA1217
|
KIAA1217 |
| chr17_+_39240459 | 0.13 |
ENST00000391417.4
|
KRTAP4-7
|
keratin associated protein 4-7 |
| chr2_+_90248739 | 0.12 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr1_+_107682629 | 0.12 |
ENST00000370074.4
ENST00000370073.2 ENST00000370071.2 ENST00000542803.1 ENST00000370061.3 ENST00000370072.3 ENST00000370070.2 |
NTNG1
|
netrin G1 |
| chr17_-_39280419 | 0.12 |
ENST00000394014.1
|
KRTAP4-12
|
keratin associated protein 4-12 |
| chr18_+_34124507 | 0.12 |
ENST00000591635.1
|
FHOD3
|
formin homology 2 domain containing 3 |
| chr8_+_76452097 | 0.12 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr13_+_77522632 | 0.12 |
ENST00000377462.1
|
IRG1
|
immunoresponsive 1 homolog (mouse) |
| chr21_-_31538971 | 0.12 |
ENST00000286808.3
|
CLDN17
|
claudin 17 |
| chr4_-_116034979 | 0.12 |
ENST00000264363.2
|
NDST4
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 |
| chr6_+_26365443 | 0.12 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr2_+_187454749 | 0.11 |
ENST00000261023.3
ENST00000374907.3 |
ITGAV
|
integrin, alpha V |
| chr11_-_30608413 | 0.11 |
ENST00000528686.1
|
MPPED2
|
metallophosphoesterase domain containing 2 |
| chr5_-_145562147 | 0.11 |
ENST00000545646.1
ENST00000274562.9 ENST00000510191.1 ENST00000394434.2 |
LARS
|
leucyl-tRNA synthetase |
| chr8_+_92261516 | 0.11 |
ENST00000276609.3
ENST00000309536.2 |
SLC26A7
|
solute carrier family 26 (anion exchanger), member 7 |
| chr17_-_77924627 | 0.11 |
ENST00000572862.1
ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16
|
TBC1 domain family, member 16 |
| chr20_+_52105495 | 0.11 |
ENST00000439873.2
|
AL354993.1
|
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr2_+_105050794 | 0.11 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr2_+_223162866 | 0.11 |
ENST00000295226.1
|
CCDC140
|
coiled-coil domain containing 140 |
| chr12_+_26348429 | 0.11 |
ENST00000242729.2
|
SSPN
|
sarcospan |
| chr7_+_50348268 | 0.11 |
ENST00000438033.1
ENST00000439701.1 |
IKZF1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr8_-_40755333 | 0.11 |
ENST00000297737.6
ENST00000315769.7 |
ZMAT4
|
zinc finger, matrin-type 4 |
| chr12_-_3862245 | 0.10 |
ENST00000252322.1
ENST00000440314.2 |
EFCAB4B
|
EF-hand calcium binding domain 4B |
| chr7_+_148287657 | 0.10 |
ENST00000307003.2
|
C7orf33
|
chromosome 7 open reading frame 33 |
| chr6_-_100912785 | 0.10 |
ENST00000369208.3
|
SIM1
|
single-minded family bHLH transcription factor 1 |
| chr4_+_88754069 | 0.10 |
ENST00000395102.4
ENST00000497649.2 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr7_-_23510086 | 0.10 |
ENST00000258729.3
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr2_-_214016314 | 0.10 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr4_-_138453559 | 0.10 |
ENST00000511115.1
|
PCDH18
|
protocadherin 18 |
| chr1_-_39347255 | 0.10 |
ENST00000454994.2
ENST00000357771.3 |
GJA9
|
gap junction protein, alpha 9, 59kDa |
| chr12_+_93096759 | 0.10 |
ENST00000544406.2
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr16_+_57279004 | 0.10 |
ENST00000219204.3
|
ARL2BP
|
ADP-ribosylation factor-like 2 binding protein |
| chr5_-_36301984 | 0.09 |
ENST00000502994.1
ENST00000515759.1 ENST00000296604.3 |
RANBP3L
|
RAN binding protein 3-like |
| chr1_-_152386732 | 0.09 |
ENST00000271835.3
|
CRNN
|
cornulin |
| chr5_+_67586465 | 0.09 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr4_+_88754113 | 0.09 |
ENST00000560249.1
ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr3_-_167191814 | 0.09 |
ENST00000466903.1
ENST00000264677.4 |
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr12_+_122688090 | 0.09 |
ENST00000324189.4
ENST00000546192.1 |
B3GNT4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr5_-_126409159 | 0.09 |
ENST00000607731.1
ENST00000535381.1 ENST00000296662.5 ENST00000509733.3 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr2_+_179318295 | 0.09 |
ENST00000442710.1
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr1_-_149900122 | 0.09 |
ENST00000271628.8
|
SF3B4
|
splicing factor 3b, subunit 4, 49kDa |
| chr18_+_32556892 | 0.08 |
ENST00000591734.1
ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr10_+_98741041 | 0.08 |
ENST00000286067.2
|
C10orf12
|
chromosome 10 open reading frame 12 |
| chr7_-_25268104 | 0.08 |
ENST00000222674.2
|
NPVF
|
neuropeptide VF precursor |
| chr8_+_22424551 | 0.08 |
ENST00000523348.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr18_+_61575200 | 0.08 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr3_-_100712292 | 0.08 |
ENST00000495063.1
ENST00000530539.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr7_-_28220354 | 0.08 |
ENST00000283928.5
|
JAZF1
|
JAZF zinc finger 1 |
| chr7_-_83278322 | 0.08 |
ENST00000307792.3
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr6_+_151042224 | 0.08 |
ENST00000358517.2
|
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr9_+_82187630 | 0.08 |
ENST00000265284.6
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr15_+_69857515 | 0.08 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr6_+_12007897 | 0.08 |
ENST00000437559.1
|
RP11-456H18.2
|
RP11-456H18.2 |
| chr9_+_82187487 | 0.08 |
ENST00000435650.1
ENST00000414465.1 ENST00000376537.4 ENST00000376534.4 |
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr8_+_99129513 | 0.08 |
ENST00000522319.1
ENST00000401707.2 |
POP1
|
processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) |
| chrX_+_78003204 | 0.08 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr5_+_126984710 | 0.08 |
ENST00000379445.3
|
CTXN3
|
cortexin 3 |
| chr4_-_87028478 | 0.07 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr17_-_40337470 | 0.07 |
ENST00000293330.1
|
HCRT
|
hypocretin (orexin) neuropeptide precursor |
| chr5_+_140855495 | 0.07 |
ENST00000308177.3
|
PCDHGC3
|
protocadherin gamma subfamily C, 3 |
| chr3_-_12587055 | 0.07 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chr2_-_207024233 | 0.07 |
ENST00000423725.1
ENST00000233190.6 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr3_+_111718173 | 0.07 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr2_-_207024134 | 0.07 |
ENST00000457011.1
ENST00000440274.1 ENST00000432169.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr19_+_10397648 | 0.07 |
ENST00000340992.4
ENST00000393717.2 |
ICAM4
|
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chr18_-_3219847 | 0.07 |
ENST00000261606.7
|
MYOM1
|
myomesin 1 |
| chr4_-_20985632 | 0.07 |
ENST00000359001.5
|
KCNIP4
|
Kv channel interacting protein 4 |
| chr2_-_227050079 | 0.07 |
ENST00000423838.1
|
AC068138.1
|
AC068138.1 |
| chr9_-_131486367 | 0.07 |
ENST00000372663.4
ENST00000406904.2 ENST00000452105.1 ENST00000372672.2 ENST00000372667.5 |
ZDHHC12
|
zinc finger, DHHC-type containing 12 |
| chr22_+_39101728 | 0.07 |
ENST00000216044.5
ENST00000484657.1 |
GTPBP1
|
GTP binding protein 1 |
| chr12_-_91573132 | 0.06 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr3_-_52090461 | 0.06 |
ENST00000296483.6
ENST00000495880.1 |
DUSP7
|
dual specificity phosphatase 7 |
| chr22_-_40929812 | 0.06 |
ENST00000422851.1
|
MKL1
|
megakaryoblastic leukemia (translocation) 1 |
| chr14_-_57272366 | 0.06 |
ENST00000554788.1
ENST00000554845.1 ENST00000408990.3 |
OTX2
|
orthodenticle homeobox 2 |
| chr4_+_37962018 | 0.06 |
ENST00000504686.1
|
PTTG2
|
pituitary tumor-transforming 2 |
| chr15_-_31393910 | 0.06 |
ENST00000397795.2
ENST00000256552.6 ENST00000559179.1 |
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr3_+_141106643 | 0.06 |
ENST00000514251.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chrX_+_139791917 | 0.06 |
ENST00000607004.1
ENST00000370535.3 |
LINC00632
|
long intergenic non-protein coding RNA 632 |
| chr3_-_138048682 | 0.06 |
ENST00000383180.2
|
NME9
|
NME/NM23 family member 9 |
| chr14_-_38064198 | 0.06 |
ENST00000250448.2
|
FOXA1
|
forkhead box A1 |
| chr1_-_67266939 | 0.06 |
ENST00000304526.2
|
INSL5
|
insulin-like 5 |
| chr15_-_75748115 | 0.06 |
ENST00000360439.4
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr2_+_45168875 | 0.06 |
ENST00000260653.3
|
SIX3
|
SIX homeobox 3 |
| chr2_-_207023918 | 0.06 |
ENST00000455934.2
ENST00000449699.1 ENST00000454195.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr1_+_154229547 | 0.06 |
ENST00000428595.1
|
UBAP2L
|
ubiquitin associated protein 2-like |
| chr9_+_82186872 | 0.06 |
ENST00000376544.3
ENST00000376520.4 |
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr12_-_16759711 | 0.06 |
ENST00000447609.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr17_-_10421853 | 0.06 |
ENST00000226207.5
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult |
| chr8_-_18744528 | 0.06 |
ENST00000523619.1
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr6_+_135502501 | 0.06 |
ENST00000527615.1
ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr1_-_185597619 | 0.05 |
ENST00000608417.1
ENST00000436955.1 |
GS1-204I12.1
|
GS1-204I12.1 |
| chr5_-_1882858 | 0.05 |
ENST00000511126.1
ENST00000231357.2 |
IRX4
|
iroquois homeobox 4 |
| chr3_-_54962100 | 0.05 |
ENST00000273286.5
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1 |
| chr3_+_121774202 | 0.05 |
ENST00000469710.1
ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86
|
CD86 molecule |
| chr14_+_73706308 | 0.05 |
ENST00000554301.1
ENST00000555445.1 |
PAPLN
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr2_+_68961905 | 0.05 |
ENST00000295381.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr12_-_110937351 | 0.05 |
ENST00000552130.2
|
VPS29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr2_-_224467093 | 0.05 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr4_-_57547454 | 0.05 |
ENST00000556376.2
|
HOPX
|
HOP homeobox |
| chr2_-_180610767 | 0.05 |
ENST00000409343.1
|
ZNF385B
|
zinc finger protein 385B |
Network of associatons between targets according to the STRING database.
First level regulatory network of LMX1B_MNX1_RAX2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.2 | 0.6 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.4 | GO:0021836 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.1 | 1.3 | GO:1901509 | positive regulation of cAMP-mediated signaling(GO:0043950) regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 1.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.3 | GO:0061346 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.1 | 0.5 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.4 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.3 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.2 | GO:1902159 | regulation of inward rectifier potassium channel activity(GO:1901979) regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.0 | 0.1 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 | 0.3 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.7 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.6 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.3 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.3 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.0 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 0.0 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.6 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.1 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.0 | 0.1 | GO:0009946 | proximal/distal axis specification(GO:0009946) neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:1990922 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.0 | 0.3 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0045404 | negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.2 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.0 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.3 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.0 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.0 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.5 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.0 | 0.2 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 0.3 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 1.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta6 complex(GO:0034685) integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.4 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.6 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 0.4 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.1 | 0.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.3 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.5 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.8 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.7 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.0 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.2 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |