Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for MAFF_MAFG
Z-value: 0.72
Transcription factors associated with MAFF_MAFG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
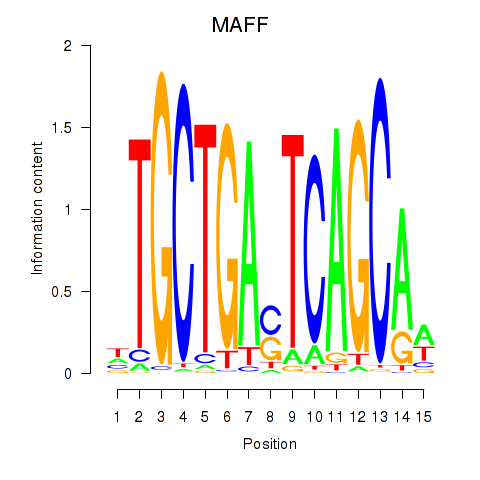
MAFF
|
ENSG00000185022.7 | MAF bZIP transcription factor F |
|
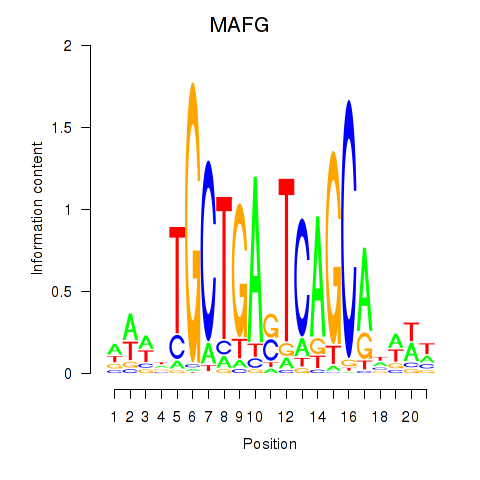
MAFG
|
ENSG00000197063.6 | MAF bZIP transcription factor G |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MAFG | hg19_v2_chr17_-_79885576_79885624 | -0.47 | 1.9e-02 | Click! |
| MAFF | hg19_v2_chr22_+_38609538_38609547 | -0.05 | 8.1e-01 | Click! |
Activity profile of MAFF_MAFG motif
Sorted Z-values of MAFF_MAFG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_132455118 | 2.46 |
ENST00000458028.1
|
LINC01013
|
long intergenic non-protein coding RNA 1013 |
| chr13_-_44453826 | 1.85 |
ENST00000444614.3
|
CCDC122
|
coiled-coil domain containing 122 |
| chr6_+_132455526 | 1.80 |
ENST00000443303.1
|
LINC01013
|
long intergenic non-protein coding RNA 1013 |
| chr16_+_31539197 | 1.64 |
ENST00000564707.1
|
AHSP
|
alpha hemoglobin stabilizing protein |
| chr8_+_9046503 | 1.64 |
ENST00000512942.2
|
RP11-10A14.5
|
RP11-10A14.5 |
| chr10_-_7513904 | 1.63 |
ENST00000420395.1
|
RP5-1031D4.2
|
RP5-1031D4.2 |
| chr2_-_86094764 | 1.54 |
ENST00000393808.3
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chrX_-_63005405 | 1.50 |
ENST00000374878.1
ENST00000437457.2 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr16_+_4838412 | 1.49 |
ENST00000589327.1
|
SMIM22
|
small integral membrane protein 22 |
| chr21_+_17909594 | 1.38 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr9_-_14693417 | 1.24 |
ENST00000380916.4
|
ZDHHC21
|
zinc finger, DHHC-type containing 21 |
| chr17_-_61512545 | 1.14 |
ENST00000585153.1
|
CYB561
|
cytochrome b561 |
| chr11_+_44748361 | 1.04 |
ENST00000533202.1
ENST00000533080.1 ENST00000520358.2 ENST00000520999.2 |
TSPAN18
|
tetraspanin 18 |
| chr10_-_116444371 | 1.00 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr5_-_111093167 | 0.99 |
ENST00000446294.2
ENST00000419114.2 |
NREP
|
neuronal regeneration related protein |
| chr17_+_32597232 | 0.91 |
ENST00000378569.2
ENST00000200307.4 ENST00000394627.1 ENST00000394630.3 |
CCL7
|
chemokine (C-C motif) ligand 7 |
| chr7_+_302918 | 0.84 |
ENST00000599994.1
|
AC187652.1
|
Protein LOC100996433 |
| chr5_-_111092873 | 0.84 |
ENST00000509025.1
ENST00000515855.1 |
NREP
|
neuronal regeneration related protein |
| chrX_-_55187531 | 0.82 |
ENST00000489298.1
ENST00000477847.2 |
FAM104B
|
family with sequence similarity 104, member B |
| chr1_+_89829610 | 0.81 |
ENST00000370456.4
ENST00000535065.1 |
GBP6
|
guanylate binding protein family, member 6 |
| chr1_-_201123546 | 0.81 |
ENST00000435310.1
ENST00000485839.2 ENST00000367330.1 |
TMEM9
|
transmembrane protein 9 |
| chr7_+_1094921 | 0.77 |
ENST00000397095.1
|
GPR146
|
G protein-coupled receptor 146 |
| chr12_+_101988627 | 0.77 |
ENST00000547405.1
ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr5_+_139027877 | 0.76 |
ENST00000302517.3
|
CXXC5
|
CXXC finger protein 5 |
| chr10_+_116853091 | 0.76 |
ENST00000526946.1
|
ATRNL1
|
attractin-like 1 |
| chr12_+_32655048 | 0.71 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chrX_+_138612889 | 0.71 |
ENST00000218099.2
ENST00000394090.2 |
F9
|
coagulation factor IX |
| chr5_-_111093406 | 0.68 |
ENST00000379671.3
|
NREP
|
neuronal regeneration related protein |
| chr9_+_71939488 | 0.68 |
ENST00000455972.1
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr11_-_114271139 | 0.67 |
ENST00000325636.4
|
C11orf71
|
chromosome 11 open reading frame 71 |
| chr17_-_63822563 | 0.65 |
ENST00000317442.8
|
CEP112
|
centrosomal protein 112kDa |
| chr5_-_111093081 | 0.65 |
ENST00000453526.2
ENST00000509427.1 |
NREP
|
neuronal regeneration related protein |
| chr8_-_91657740 | 0.63 |
ENST00000422900.1
|
TMEM64
|
transmembrane protein 64 |
| chr3_-_142720267 | 0.62 |
ENST00000597953.1
|
RP11-91G21.1
|
RP11-91G21.1 |
| chr18_-_53069419 | 0.60 |
ENST00000570177.2
|
TCF4
|
transcription factor 4 |
| chr18_-_53068911 | 0.60 |
ENST00000537856.3
|
TCF4
|
transcription factor 4 |
| chr21_-_27423339 | 0.59 |
ENST00000415997.1
|
APP
|
amyloid beta (A4) precursor protein |
| chrX_-_10544942 | 0.58 |
ENST00000380779.1
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr9_-_113100088 | 0.57 |
ENST00000374510.4
ENST00000423740.2 ENST00000374511.3 ENST00000374507.4 |
TXNDC8
|
thioredoxin domain containing 8 (spermatozoa) |
| chr17_+_60758814 | 0.57 |
ENST00000579432.1
ENST00000446119.2 |
MRC2
|
mannose receptor, C type 2 |
| chr22_-_27014043 | 0.57 |
ENST00000215939.2
|
CRYBB1
|
crystallin, beta B1 |
| chr6_+_33043703 | 0.57 |
ENST00000418931.2
ENST00000535465.1 |
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chr12_+_101988774 | 0.56 |
ENST00000545503.2
ENST00000536007.1 ENST00000541119.1 ENST00000361466.2 ENST00000551300.1 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chrX_-_55187588 | 0.56 |
ENST00000472571.2
ENST00000332132.4 ENST00000425133.2 ENST00000358460.4 |
FAM104B
|
family with sequence similarity 104, member B |
| chr12_+_51318513 | 0.56 |
ENST00000332160.4
|
METTL7A
|
methyltransferase like 7A |
| chr22_+_23089870 | 0.55 |
ENST00000390311.2
|
IGLV3-16
|
immunoglobulin lambda variable 3-16 |
| chr16_-_24942273 | 0.54 |
ENST00000571406.1
|
ARHGAP17
|
Rho GTPase activating protein 17 |
| chr1_+_196788887 | 0.53 |
ENST00000320493.5
ENST00000367424.4 ENST00000367421.3 |
CFHR1
CFHR2
|
complement factor H-related 1 complement factor H-related 2 |
| chr6_+_80816342 | 0.53 |
ENST00000369760.4
ENST00000356489.5 ENST00000320393.6 |
BCKDHB
|
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chr6_-_90529418 | 0.53 |
ENST00000439638.1
ENST00000369393.3 ENST00000428876.1 |
MDN1
|
MDN1, midasin homolog (yeast) |
| chr9_-_34376851 | 0.52 |
ENST00000297625.7
|
KIAA1161
|
KIAA1161 |
| chr7_+_100663353 | 0.52 |
ENST00000306151.4
|
MUC17
|
mucin 17, cell surface associated |
| chr4_+_26585538 | 0.52 |
ENST00000264866.4
|
TBC1D19
|
TBC1 domain family, member 19 |
| chr19_-_10491130 | 0.51 |
ENST00000530829.1
ENST00000529370.1 |
TYK2
|
tyrosine kinase 2 |
| chr4_-_123843597 | 0.51 |
ENST00000510735.1
ENST00000304430.5 |
NUDT6
|
nudix (nucleoside diphosphate linked moiety X)-type motif 6 |
| chr5_-_111093340 | 0.50 |
ENST00000508870.1
|
NREP
|
neuronal regeneration related protein |
| chr4_-_89080003 | 0.50 |
ENST00000237612.3
|
ABCG2
|
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr8_+_96037255 | 0.50 |
ENST00000286687.4
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr5_+_176449684 | 0.49 |
ENST00000506693.1
ENST00000358149.3 ENST00000512315.1 ENST00000503425.1 |
ZNF346
|
zinc finger protein 346 |
| chr20_-_56195449 | 0.49 |
ENST00000541799.1
|
ZBP1
|
Z-DNA binding protein 1 |
| chr17_+_66509019 | 0.49 |
ENST00000585981.1
ENST00000589480.1 ENST00000585815.1 |
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chrX_-_119709637 | 0.48 |
ENST00000404115.3
|
CUL4B
|
cullin 4B |
| chr11_+_121447469 | 0.48 |
ENST00000532694.1
ENST00000534286.1 |
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr4_+_26585686 | 0.45 |
ENST00000505206.1
ENST00000511789.1 |
TBC1D19
|
TBC1 domain family, member 19 |
| chr22_+_23154239 | 0.44 |
ENST00000390315.2
|
IGLV3-10
|
immunoglobulin lambda variable 3-10 |
| chr7_+_139025875 | 0.44 |
ENST00000297534.6
|
C7orf55
|
chromosome 7 open reading frame 55 |
| chr17_-_14140166 | 0.43 |
ENST00000420162.2
ENST00000431716.2 |
CDRT15
|
CMT1A duplicated region transcript 15 |
| chr1_+_2004901 | 0.42 |
ENST00000400921.2
|
PRKCZ
|
protein kinase C, zeta |
| chr17_-_34313685 | 0.42 |
ENST00000435911.2
ENST00000586216.1 ENST00000394509.4 |
CCL14
|
chemokine (C-C motif) ligand 14 |
| chr1_+_196912902 | 0.41 |
ENST00000476712.2
ENST00000367415.5 |
CFHR2
|
complement factor H-related 2 |
| chr1_+_17248418 | 0.41 |
ENST00000375541.5
|
CROCC
|
ciliary rootlet coiled-coil, rootletin |
| chr4_-_42659102 | 0.40 |
ENST00000264449.10
ENST00000510289.1 ENST00000381668.5 |
ATP8A1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 |
| chr22_+_22730353 | 0.40 |
ENST00000390296.2
|
IGLV5-45
|
immunoglobulin lambda variable 5-45 |
| chr5_+_140579162 | 0.40 |
ENST00000536699.1
ENST00000354757.3 |
PCDHB11
|
protocadherin beta 11 |
| chr19_+_10736183 | 0.38 |
ENST00000590857.1
ENST00000588688.1 ENST00000586078.1 |
SLC44A2
|
solute carrier family 44 (choline transporter), member 2 |
| chr12_+_10365404 | 0.38 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr16_+_4838393 | 0.38 |
ENST00000589721.1
|
SMIM22
|
small integral membrane protein 22 |
| chr5_-_42811986 | 0.38 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr19_-_23456996 | 0.38 |
ENST00000594653.1
|
RP11-15H20.7
|
RP11-15H20.7 |
| chr7_-_17980091 | 0.37 |
ENST00000409389.1
ENST00000409604.1 ENST00000428135.3 |
SNX13
|
sorting nexin 13 |
| chr14_-_75530693 | 0.37 |
ENST00000555135.1
ENST00000357971.3 ENST00000553302.1 ENST00000555694.1 ENST00000238618.3 |
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr19_+_16254488 | 0.37 |
ENST00000588246.1
ENST00000593031.1 |
HSH2D
|
hematopoietic SH2 domain containing |
| chr6_+_150070857 | 0.37 |
ENST00000544496.1
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr11_-_119211525 | 0.36 |
ENST00000528368.1
|
C1QTNF5
|
C1q and tumor necrosis factor related protein 5 |
| chr9_-_34710066 | 0.36 |
ENST00000378792.1
ENST00000259607.2 |
CCL21
|
chemokine (C-C motif) ligand 21 |
| chr1_+_179051160 | 0.36 |
ENST00000367625.4
ENST00000352445.6 |
TOR3A
|
torsin family 3, member A |
| chr2_+_87808725 | 0.35 |
ENST00000413202.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr1_-_201123586 | 0.35 |
ENST00000414605.2
ENST00000367334.5 ENST00000367332.1 |
TMEM9
|
transmembrane protein 9 |
| chr17_+_18625336 | 0.34 |
ENST00000395671.4
ENST00000571542.1 ENST00000395672.2 ENST00000414850.2 ENST00000424146.2 |
TRIM16L
|
tripartite motif containing 16-like |
| chr3_+_45071622 | 0.34 |
ENST00000428034.1
|
CLEC3B
|
C-type lectin domain family 3, member B |
| chr8_-_135522425 | 0.34 |
ENST00000521673.1
|
ZFAT
|
zinc finger and AT hook domain containing |
| chr19_-_14785698 | 0.34 |
ENST00000344373.4
ENST00000595472.1 |
EMR3
|
egf-like module containing, mucin-like, hormone receptor-like 3 |
| chr12_+_45609797 | 0.33 |
ENST00000425752.2
|
ANO6
|
anoctamin 6 |
| chr8_+_96037205 | 0.33 |
ENST00000396124.4
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr5_+_140602904 | 0.33 |
ENST00000515856.2
ENST00000239449.4 |
PCDHB14
|
protocadherin beta 14 |
| chr2_+_115219171 | 0.33 |
ENST00000409163.1
|
DPP10
|
dipeptidyl-peptidase 10 (non-functional) |
| chr7_-_30518387 | 0.32 |
ENST00000222823.4
ENST00000419601.1 |
NOD1
|
nucleotide-binding oligomerization domain containing 1 |
| chr18_-_52989217 | 0.32 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr10_-_100174900 | 0.32 |
ENST00000370575.4
|
PYROXD2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr11_-_914682 | 0.32 |
ENST00000323541.7
|
CHID1
|
chitinase domain containing 1 |
| chrX_+_102883887 | 0.32 |
ENST00000372625.3
ENST00000372624.3 |
TCEAL1
|
transcription elongation factor A (SII)-like 1 |
| chr21_-_27543425 | 0.32 |
ENST00000448388.2
|
APP
|
amyloid beta (A4) precursor protein |
| chr5_-_42812143 | 0.31 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr11_-_77791156 | 0.31 |
ENST00000281031.4
|
NDUFC2
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa |
| chr3_-_125802765 | 0.31 |
ENST00000514891.1
ENST00000512470.1 ENST00000504035.1 ENST00000360370.4 ENST00000513723.1 ENST00000510651.1 ENST00000514333.1 |
SLC41A3
|
solute carrier family 41, member 3 |
| chr15_-_20170354 | 0.30 |
ENST00000338912.5
|
IGHV1OR15-9
|
immunoglobulin heavy variable 1/OR15-9 (non-functional) |
| chr1_+_52682052 | 0.29 |
ENST00000371591.1
|
ZFYVE9
|
zinc finger, FYVE domain containing 9 |
| chr6_+_27215494 | 0.29 |
ENST00000230582.3
|
PRSS16
|
protease, serine, 16 (thymus) |
| chr22_+_22550113 | 0.29 |
ENST00000390285.3
|
IGLV6-57
|
immunoglobulin lambda variable 6-57 |
| chr18_-_52989525 | 0.29 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr5_-_149535421 | 0.29 |
ENST00000261799.4
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide |
| chr17_-_76899275 | 0.29 |
ENST00000322630.2
ENST00000586713.1 |
DDC8
|
Protein DDC8 homolog |
| chr5_+_176449726 | 0.29 |
ENST00000261948.4
ENST00000511834.1 ENST00000503039.1 |
ZNF346
|
zinc finger protein 346 |
| chr17_+_20483037 | 0.29 |
ENST00000399044.1
|
CDRT15L2
|
CMT1A duplicated region transcript 15-like 2 |
| chr17_-_76921459 | 0.28 |
ENST00000262768.7
|
TIMP2
|
TIMP metallopeptidase inhibitor 2 |
| chr18_-_53257027 | 0.27 |
ENST00000568740.1
ENST00000564403.2 ENST00000537578.1 |
TCF4
|
transcription factor 4 |
| chr17_-_79166176 | 0.27 |
ENST00000571292.1
|
AZI1
|
5-azacytidine induced 1 |
| chr16_-_23521710 | 0.27 |
ENST00000562117.1
ENST00000567468.1 ENST00000562944.1 ENST00000309859.4 |
GGA2
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 2 |
| chr2_-_46844159 | 0.27 |
ENST00000474980.1
ENST00000306465.4 |
PIGF
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr6_-_42946888 | 0.27 |
ENST00000244546.4
|
PEX6
|
peroxisomal biogenesis factor 6 |
| chr19_+_5914213 | 0.27 |
ENST00000222125.5
ENST00000452990.2 ENST00000588865.1 |
CAPS
|
calcyphosine |
| chr7_+_107220660 | 0.27 |
ENST00000465919.1
ENST00000445771.2 ENST00000479917.1 ENST00000421217.1 ENST00000457837.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chr11_+_67171358 | 0.26 |
ENST00000526387.1
|
TBC1D10C
|
TBC1 domain family, member 10C |
| chr17_-_64187973 | 0.26 |
ENST00000583358.1
ENST00000392769.2 |
CEP112
|
centrosomal protein 112kDa |
| chr5_-_115152651 | 0.26 |
ENST00000250535.4
|
CDO1
|
cysteine dioxygenase type 1 |
| chr15_+_34517194 | 0.26 |
ENST00000267750.4
ENST00000249209.4 ENST00000561372.1 ENST00000559078.1 ENST00000557879.1 |
EMC4
|
ER membrane protein complex subunit 4 |
| chr6_+_30951487 | 0.26 |
ENST00000486149.2
ENST00000376296.3 |
MUC21
|
mucin 21, cell surface associated |
| chr14_-_72458326 | 0.26 |
ENST00000542853.1
|
AC005477.1
|
AC005477.1 |
| chr15_+_58724184 | 0.26 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr6_+_150070831 | 0.25 |
ENST00000367380.5
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr8_+_94712732 | 0.25 |
ENST00000518322.1
|
FAM92A1
|
family with sequence similarity 92, member A1 |
| chr12_-_71533055 | 0.25 |
ENST00000552128.1
|
TSPAN8
|
tetraspanin 8 |
| chr11_-_59578202 | 0.25 |
ENST00000300151.4
|
MRPL16
|
mitochondrial ribosomal protein L16 |
| chr1_+_110254850 | 0.25 |
ENST00000369812.5
ENST00000256593.3 ENST00000369813.1 |
GSTM5
|
glutathione S-transferase mu 5 |
| chr16_+_82068830 | 0.25 |
ENST00000199936.4
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr2_-_46844242 | 0.25 |
ENST00000281382.6
|
PIGF
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr12_+_111051902 | 0.25 |
ENST00000397655.3
ENST00000471804.2 ENST00000377654.3 ENST00000397659.4 |
TCTN1
|
tectonic family member 1 |
| chr11_+_67171391 | 0.25 |
ENST00000312390.5
|
TBC1D10C
|
TBC1 domain family, member 10C |
| chr22_+_23213658 | 0.24 |
ENST00000390318.2
|
IGLV4-3
|
immunoglobulin lambda variable 4-3 |
| chr2_+_74781828 | 0.24 |
ENST00000340004.6
|
DOK1
|
docking protein 1, 62kDa (downstream of tyrosine kinase 1) |
| chr10_-_48416849 | 0.24 |
ENST00000249598.1
|
GDF2
|
growth differentiation factor 2 |
| chr6_-_167571817 | 0.24 |
ENST00000366834.1
|
GPR31
|
G protein-coupled receptor 31 |
| chr2_+_46844290 | 0.24 |
ENST00000238892.3
|
CRIPT
|
cysteine-rich PDZ-binding protein |
| chr16_-_18801643 | 0.24 |
ENST00000322989.4
ENST00000563390.1 |
RPS15A
|
ribosomal protein S15a |
| chr10_+_5488564 | 0.23 |
ENST00000449083.1
ENST00000380359.3 |
NET1
|
neuroepithelial cell transforming 1 |
| chr3_+_111451321 | 0.23 |
ENST00000393923.3
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr12_-_102455846 | 0.23 |
ENST00000545679.1
|
CCDC53
|
coiled-coil domain containing 53 |
| chr1_-_23694794 | 0.23 |
ENST00000374608.3
|
ZNF436
|
zinc finger protein 436 |
| chr16_+_640201 | 0.23 |
ENST00000563109.1
|
RAB40C
|
RAB40C, member RAS oncogene family |
| chr5_-_54468974 | 0.23 |
ENST00000381375.2
ENST00000296733.1 ENST00000322374.6 ENST00000334206.5 ENST00000331730.3 |
CDC20B
|
cell division cycle 20B |
| chr5_-_66492562 | 0.23 |
ENST00000256447.4
|
CD180
|
CD180 molecule |
| chr17_+_66521936 | 0.23 |
ENST00000592800.1
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr1_+_22979676 | 0.23 |
ENST00000432749.2
ENST00000314933.6 |
C1QB
|
complement component 1, q subcomponent, B chain |
| chr4_+_15480828 | 0.22 |
ENST00000389652.5
|
CC2D2A
|
coiled-coil and C2 domain containing 2A |
| chr11_+_72983246 | 0.22 |
ENST00000393590.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr20_+_61867235 | 0.22 |
ENST00000342412.6
ENST00000217169.3 |
BIRC7
|
baculoviral IAP repeat containing 7 |
| chr17_+_66508154 | 0.22 |
ENST00000358598.2
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr17_-_3496171 | 0.22 |
ENST00000399756.4
|
TRPV1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr5_+_99871004 | 0.22 |
ENST00000312637.4
|
FAM174A
|
family with sequence similarity 174, member A |
| chr3_-_178969403 | 0.21 |
ENST00000314235.5
ENST00000392685.2 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr11_-_94965667 | 0.21 |
ENST00000542176.1
ENST00000278499.2 |
SESN3
|
sestrin 3 |
| chr7_+_75911902 | 0.21 |
ENST00000413003.1
|
SRRM3
|
serine/arginine repetitive matrix 3 |
| chr5_-_55412774 | 0.21 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr1_-_229644034 | 0.21 |
ENST00000366678.3
ENST00000261396.3 ENST00000537506.1 |
NUP133
|
nucleoporin 133kDa |
| chr1_+_24104869 | 0.21 |
ENST00000246151.4
|
PITHD1
|
PITH (C-terminal proteasome-interacting domain of thioredoxin-like) domain containing 1 |
| chr1_-_160313025 | 0.21 |
ENST00000368069.3
ENST00000241704.7 |
COPA
|
coatomer protein complex, subunit alpha |
| chr3_-_3221358 | 0.20 |
ENST00000424814.1
ENST00000450014.1 ENST00000231948.4 ENST00000432408.2 |
CRBN
|
cereblon |
| chr14_+_58894103 | 0.20 |
ENST00000354386.6
ENST00000556134.1 |
KIAA0586
|
KIAA0586 |
| chr12_+_105501487 | 0.20 |
ENST00000332180.5
|
KIAA1033
|
KIAA1033 |
| chr14_-_106733624 | 0.19 |
ENST00000390610.2
|
IGHV1-24
|
immunoglobulin heavy variable 1-24 |
| chr4_+_147560042 | 0.19 |
ENST00000281321.3
|
POU4F2
|
POU class 4 homeobox 2 |
| chr17_-_28618948 | 0.19 |
ENST00000261714.6
|
BLMH
|
bleomycin hydrolase |
| chr11_-_104769141 | 0.19 |
ENST00000508062.1
ENST00000422698.2 |
CASP12
|
caspase 12 (gene/pseudogene) |
| chr11_-_915012 | 0.19 |
ENST00000449825.1
ENST00000533056.1 |
CHID1
|
chitinase domain containing 1 |
| chr7_-_99679324 | 0.19 |
ENST00000292393.5
ENST00000413658.2 ENST00000412947.1 ENST00000441298.1 ENST00000449785.1 ENST00000299667.4 ENST00000424697.1 |
ZNF3
|
zinc finger protein 3 |
| chr17_+_54230819 | 0.19 |
ENST00000318698.2
ENST00000566473.2 |
ANKFN1
|
ankyrin-repeat and fibronectin type III domain containing 1 |
| chrX_+_129473916 | 0.18 |
ENST00000545805.1
ENST00000543953.1 ENST00000218197.5 |
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr13_-_53024725 | 0.18 |
ENST00000378060.4
|
VPS36
|
vacuolar protein sorting 36 homolog (S. cerevisiae) |
| chrX_-_133931164 | 0.18 |
ENST00000370790.1
ENST00000298090.6 |
FAM122B
|
family with sequence similarity 122B |
| chr17_-_28618867 | 0.18 |
ENST00000394819.3
ENST00000577623.1 |
BLMH
|
bleomycin hydrolase |
| chr17_-_61920280 | 0.18 |
ENST00000448276.2
ENST00000577990.1 |
SMARCD2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 |
| chr2_+_201981119 | 0.18 |
ENST00000395148.2
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr6_-_159420780 | 0.18 |
ENST00000449822.1
|
RSPH3
|
radial spoke 3 homolog (Chlamydomonas) |
| chr8_+_64081118 | 0.18 |
ENST00000539294.1
|
YTHDF3
|
YTH domain family, member 3 |
| chr6_+_29364416 | 0.18 |
ENST00000383555.2
|
OR12D2
|
olfactory receptor, family 12, subfamily D, member 2 (gene/pseudogene) |
| chr1_-_166845515 | 0.18 |
ENST00000367874.4
|
TADA1
|
transcriptional adaptor 1 |
| chrX_-_64196351 | 0.17 |
ENST00000374839.3
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr10_-_14590644 | 0.17 |
ENST00000378470.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr16_-_24942655 | 0.17 |
ENST00000573765.1
|
ARHGAP17
|
Rho GTPase activating protein 17 |
| chr3_-_46506358 | 0.17 |
ENST00000417439.1
ENST00000431944.1 |
LTF
|
lactotransferrin |
| chr2_-_187713891 | 0.17 |
ENST00000295131.2
|
ZSWIM2
|
zinc finger, SWIM-type containing 2 |
| chr14_+_58894141 | 0.17 |
ENST00000423743.3
|
KIAA0586
|
KIAA0586 |
| chr7_+_44040488 | 0.17 |
ENST00000258704.3
|
SPDYE1
|
speedy/RINGO cell cycle regulator family member E1 |
| chr4_-_140222358 | 0.16 |
ENST00000505036.1
ENST00000544855.1 ENST00000539002.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr7_-_86849883 | 0.16 |
ENST00000433078.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr16_+_31539183 | 0.16 |
ENST00000302312.4
|
AHSP
|
alpha hemoglobin stabilizing protein |
| chr12_+_45609862 | 0.16 |
ENST00000423947.3
ENST00000435642.1 |
ANO6
|
anoctamin 6 |
| chr9_+_134378289 | 0.16 |
ENST00000423007.1
ENST00000404875.2 ENST00000441334.1 ENST00000341012.7 ENST00000372228.3 ENST00000402686.3 ENST00000419118.2 ENST00000541219.1 ENST00000354713.4 ENST00000418774.1 ENST00000415075.1 ENST00000448212.1 ENST00000430619.1 |
POMT1
|
protein-O-mannosyltransferase 1 |
| chr11_-_34938039 | 0.16 |
ENST00000395787.3
|
APIP
|
APAF1 interacting protein |
| chrX_-_64196307 | 0.16 |
ENST00000545618.1
|
ZC4H2
|
zinc finger, C4H2 domain containing |
Network of associatons between targets according to the STRING database.
First level regulatory network of MAFF_MAFG
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.2 | 0.5 | GO:1902948 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 | 0.9 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 1.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) sebaceous gland development(GO:0048733) |
| 0.1 | 0.5 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.1 | 0.4 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.1 | 1.5 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.6 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 0.3 | GO:0033214 | iron assimilation(GO:0033212) iron assimilation by chelation and transport(GO:0033214) positive regulation of bone mineralization involved in bone maturation(GO:1900159) negative regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000308) |
| 0.1 | 0.4 | GO:1903566 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.4 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 | 0.1 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 0.3 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.1 | 1.8 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.1 | 0.7 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 1.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.3 | GO:0072275 | cell migration involved in vasculogenesis(GO:0035441) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 0.2 | GO:0050960 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.1 | 0.5 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 1.1 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.5 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 0.3 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.1 | 0.3 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.1 | 0.5 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.4 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.1 | 0.1 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.1 | 0.8 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.6 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.3 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.1 | 0.6 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 0.2 | GO:0051664 | nuclear pore distribution(GO:0031081) nuclear pore localization(GO:0051664) |
| 0.0 | 0.3 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.2 | GO:1902869 | regulation of amacrine cell differentiation(GO:1902869) |
| 0.0 | 0.1 | GO:1900081 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.1 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.0 | 0.1 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.2 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.6 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.2 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.1 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.2 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.0 | 0.1 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.6 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.5 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.0 | 0.1 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.2 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.1 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.0 | 0.1 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.0 | 0.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.9 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.5 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.4 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.0 | 0.1 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.0 | 0.0 | GO:0036446 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 | 0.3 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.4 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.4 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.4 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.1 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.3 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 1.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.2 | GO:0070269 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) pyroptosis(GO:0070269) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 | 0.3 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.1 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 1.3 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 1.1 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.1 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.0 | 0.3 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:0019477 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 0.0 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.5 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.1 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.0 | 0.2 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.4 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.2 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.0 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.3 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.9 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.1 | 0.5 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.4 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.1 | 0.2 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 0.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.3 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 1.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.3 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.0 | 0.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.4 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.5 | GO:0045179 | axon hillock(GO:0043203) apical cortex(GO:0045179) |
| 0.0 | 1.1 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.1 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.5 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.6 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 1.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.3 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.3 | GO:0042599 | lamellar body(GO:0042599) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.3 | 1.1 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.2 | 0.5 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.2 | 2.1 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.5 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.9 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 1.5 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.6 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.5 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.9 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 0.3 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.1 | 0.3 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.1 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 0.4 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 1.3 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 0.5 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 1.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.3 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.1 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.3 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.4 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.6 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 1.2 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.7 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.1 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.5 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.8 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.5 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.5 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0000035 | acyl binding(GO:0000035) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.4 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.0 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.1 | GO:0002060 | purine nucleobase binding(GO:0002060) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.0 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.0 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.5 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.4 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 1.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.3 | PID S1P S1P1 PATHWAY | S1P1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.7 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 1.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 2.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 1.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 1.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.6 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.3 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |