Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
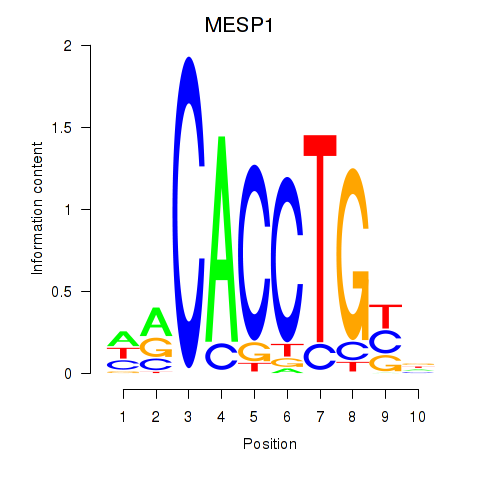
Results for MESP1
Z-value: 0.87
Transcription factors associated with MESP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MESP1
|
ENSG00000166823.5 | mesoderm posterior bHLH transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MESP1 | hg19_v2_chr15_-_90294523_90294541 | 0.13 | 5.5e-01 | Click! |
Activity profile of MESP1 motif
Sorted Z-values of MESP1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_39382900 | 9.59 |
ENST00000377721.3
ENST00000455970.2 |
KRTAP9-2
|
keratin associated protein 9-2 |
| chr17_+_39394250 | 4.21 |
ENST00000254072.6
|
KRTAP9-8
|
keratin associated protein 9-8 |
| chr10_+_94608218 | 2.69 |
ENST00000371543.1
|
EXOC6
|
exocyst complex component 6 |
| chr2_+_241631262 | 1.48 |
ENST00000337801.4
ENST00000429564.1 |
AQP12A
|
aquaporin 12A |
| chr2_-_241622278 | 1.45 |
ENST00000407834.3
|
AQP12B
|
aquaporin 12B |
| chr14_+_75746340 | 1.33 |
ENST00000555686.1
ENST00000555672.1 |
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr2_-_157189180 | 1.33 |
ENST00000539077.1
ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr4_+_156588806 | 1.27 |
ENST00000513574.1
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr14_+_75746781 | 1.20 |
ENST00000555347.1
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr8_-_93029865 | 1.19 |
ENST00000422361.2
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr20_-_39317868 | 1.17 |
ENST00000373313.2
|
MAFB
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B |
| chr1_-_209979465 | 1.15 |
ENST00000542854.1
|
IRF6
|
interferon regulatory factor 6 |
| chr13_+_31480328 | 1.10 |
ENST00000380482.4
|
MEDAG
|
mesenteric estrogen-dependent adipogenesis |
| chr8_+_120220561 | 1.09 |
ENST00000276681.6
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene) |
| chr3_-_49459865 | 1.08 |
ENST00000427987.1
|
AMT
|
aminomethyltransferase |
| chr3_-_49459878 | 1.06 |
ENST00000546031.1
ENST00000458307.2 ENST00000430521.1 |
AMT
|
aminomethyltransferase |
| chr12_-_42877726 | 1.06 |
ENST00000548696.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr10_-_93392811 | 1.05 |
ENST00000238994.5
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C |
| chr12_-_42877764 | 1.04 |
ENST00000455697.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr11_-_33913708 | 0.93 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr1_-_209979375 | 0.91 |
ENST00000367021.3
|
IRF6
|
interferon regulatory factor 6 |
| chr3_+_43328004 | 0.90 |
ENST00000454177.1
ENST00000429705.2 ENST00000296088.7 ENST00000437827.1 |
SNRK
|
SNF related kinase |
| chr19_+_48673949 | 0.90 |
ENST00000328759.7
|
C19orf68
|
chromosome 19 open reading frame 68 |
| chr6_+_18387570 | 0.89 |
ENST00000259939.3
|
RNF144B
|
ring finger protein 144B |
| chr1_+_61542922 | 0.88 |
ENST00000407417.3
|
NFIA
|
nuclear factor I/A |
| chr1_+_26438289 | 0.87 |
ENST00000374271.4
ENST00000374269.1 |
PDIK1L
|
PDLIM1 interacting kinase 1 like |
| chr3_-_128294929 | 0.87 |
ENST00000356020.2
|
C3orf27
|
chromosome 3 open reading frame 27 |
| chr4_+_156588350 | 0.87 |
ENST00000296518.7
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr7_+_12727250 | 0.85 |
ENST00000404894.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr4_+_156588249 | 0.84 |
ENST00000393832.3
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr2_+_219745020 | 0.83 |
ENST00000258411.3
|
WNT10A
|
wingless-type MMTV integration site family, member 10A |
| chr20_-_23030296 | 0.83 |
ENST00000377103.2
|
THBD
|
thrombomodulin |
| chr4_+_156588115 | 0.83 |
ENST00000455639.2
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr12_+_32655048 | 0.82 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr5_+_68788594 | 0.82 |
ENST00000396442.2
ENST00000380766.2 |
OCLN
|
occludin |
| chr1_-_45672221 | 0.82 |
ENST00000359600.5
|
ZSWIM5
|
zinc finger, SWIM-type containing 5 |
| chr17_+_73521763 | 0.82 |
ENST00000167462.5
ENST00000375227.4 ENST00000392550.3 ENST00000578363.1 ENST00000579392.1 |
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr5_-_137071756 | 0.81 |
ENST00000394937.3
ENST00000309755.4 |
KLHL3
|
kelch-like family member 3 |
| chr4_+_156587979 | 0.81 |
ENST00000511507.1
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr7_+_22766766 | 0.78 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr8_-_30891078 | 0.78 |
ENST00000339382.2
ENST00000475541.1 |
PURG
|
purine-rich element binding protein G |
| chr6_-_130031358 | 0.77 |
ENST00000368149.2
|
ARHGAP18
|
Rho GTPase activating protein 18 |
| chr12_-_95044309 | 0.77 |
ENST00000261226.4
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr10_+_94608245 | 0.76 |
ENST00000443748.2
ENST00000260762.6 |
EXOC6
|
exocyst complex component 6 |
| chr8_-_127570603 | 0.76 |
ENST00000304916.3
|
FAM84B
|
family with sequence similarity 84, member B |
| chr8_-_103668114 | 0.72 |
ENST00000285407.6
|
KLF10
|
Kruppel-like factor 10 |
| chr20_+_42574317 | 0.71 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2 |
| chr13_-_52980263 | 0.70 |
ENST00000258613.4
ENST00000544466.1 |
THSD1
|
thrombospondin, type I, domain containing 1 |
| chr11_+_68080077 | 0.70 |
ENST00000294304.7
|
LRP5
|
low density lipoprotein receptor-related protein 5 |
| chr2_-_238499303 | 0.70 |
ENST00000409576.1
|
RAB17
|
RAB17, member RAS oncogene family |
| chr12_-_96184533 | 0.69 |
ENST00000343702.4
ENST00000344911.4 |
NTN4
|
netrin 4 |
| chr13_-_77460525 | 0.68 |
ENST00000377474.2
ENST00000317765.2 |
KCTD12
|
potassium channel tetramerization domain containing 12 |
| chr5_+_68710906 | 0.67 |
ENST00000325631.5
ENST00000454295.2 |
MARVELD2
|
MARVEL domain containing 2 |
| chr10_-_33625154 | 0.66 |
ENST00000265371.4
|
NRP1
|
neuropilin 1 |
| chr3_+_32280159 | 0.66 |
ENST00000458535.2
ENST00000307526.3 |
CMTM8
|
CKLF-like MARVEL transmembrane domain containing 8 |
| chr15_+_75639296 | 0.65 |
ENST00000564500.1
ENST00000355059.4 ENST00000566752.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr1_+_6615241 | 0.64 |
ENST00000333172.6
ENST00000328191.4 ENST00000351136.3 |
TAS1R1
|
taste receptor, type 1, member 1 |
| chr16_+_4845379 | 0.63 |
ENST00000588606.1
ENST00000586005.1 |
SMIM22
|
small integral membrane protein 22 |
| chr15_+_75640068 | 0.62 |
ENST00000565051.1
ENST00000564257.1 ENST00000567005.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr16_-_89268070 | 0.62 |
ENST00000562855.2
|
SLC22A31
|
solute carrier family 22, member 31 |
| chr2_-_75788038 | 0.62 |
ENST00000393913.3
ENST00000410113.1 |
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr19_+_54058073 | 0.59 |
ENST00000505949.1
ENST00000513265.1 |
ZNF331
|
zinc finger protein 331 |
| chr1_-_33647267 | 0.58 |
ENST00000291416.5
|
TRIM62
|
tripartite motif containing 62 |
| chr11_-_118122996 | 0.57 |
ENST00000525386.1
ENST00000527472.1 ENST00000278949.4 |
MPZL3
|
myelin protein zero-like 3 |
| chr19_+_6464502 | 0.56 |
ENST00000308243.7
|
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr5_+_78532003 | 0.56 |
ENST00000396137.4
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr9_+_71986182 | 0.54 |
ENST00000303068.7
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr4_+_41540160 | 0.54 |
ENST00000503057.1
ENST00000511496.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr9_-_126030817 | 0.52 |
ENST00000348403.5
ENST00000447404.2 ENST00000360998.3 |
STRBP
|
spermatid perinuclear RNA binding protein |
| chr6_-_30712313 | 0.52 |
ENST00000376377.2
ENST00000259874.5 |
IER3
|
immediate early response 3 |
| chr2_-_165477971 | 0.52 |
ENST00000446413.2
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr15_+_75639951 | 0.52 |
ENST00000564784.1
ENST00000569035.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr1_+_178694300 | 0.51 |
ENST00000367635.3
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr7_+_30960915 | 0.50 |
ENST00000441328.2
ENST00000409899.1 ENST00000409611.1 |
AQP1
|
aquaporin 1 (Colton blood group) |
| chrX_+_118108601 | 0.50 |
ENST00000371628.3
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr10_-_103347883 | 0.50 |
ENST00000339310.3
ENST00000370158.3 ENST00000299206.4 ENST00000456836.2 ENST00000413344.1 ENST00000429502.1 ENST00000430045.1 ENST00000370172.1 ENST00000436284.2 ENST00000370162.3 |
POLL
|
polymerase (DNA directed), lambda |
| chr7_+_16793160 | 0.49 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr3_+_57094469 | 0.48 |
ENST00000334325.1
|
SPATA12
|
spermatogenesis associated 12 |
| chr2_+_203499901 | 0.48 |
ENST00000303116.6
ENST00000392238.2 |
FAM117B
|
family with sequence similarity 117, member B |
| chr17_+_4981535 | 0.48 |
ENST00000318833.3
|
ZFP3
|
ZFP3 zinc finger protein |
| chr17_-_21156578 | 0.47 |
ENST00000399011.2
ENST00000468196.1 |
C17orf103
|
chromosome 17 open reading frame 103 |
| chr7_+_12726474 | 0.47 |
ENST00000396662.1
ENST00000356797.3 ENST00000396664.2 |
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr19_+_7660716 | 0.47 |
ENST00000160298.4
ENST00000446248.2 |
CAMSAP3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr16_+_3162557 | 0.46 |
ENST00000382192.3
ENST00000219091.4 ENST00000444510.2 ENST00000414351.1 |
ZNF205
|
zinc finger protein 205 |
| chr19_+_42381337 | 0.46 |
ENST00000597454.1
ENST00000444740.2 |
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chrX_+_118108571 | 0.46 |
ENST00000304778.7
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr4_+_75310851 | 0.46 |
ENST00000395748.3
ENST00000264487.2 |
AREG
|
amphiregulin |
| chr9_-_116061476 | 0.45 |
ENST00000441031.3
|
RNF183
|
ring finger protein 183 |
| chr13_+_112721913 | 0.45 |
ENST00000330949.1
|
SOX1
|
SRY (sex determining region Y)-box 1 |
| chr7_-_38670957 | 0.45 |
ENST00000325590.5
ENST00000428293.2 |
AMPH
|
amphiphysin |
| chr1_+_78354175 | 0.44 |
ENST00000401035.3
ENST00000457030.1 ENST00000330010.8 |
NEXN
|
nexilin (F actin binding protein) |
| chr12_-_48398104 | 0.44 |
ENST00000337299.6
ENST00000380518.3 |
COL2A1
|
collagen, type II, alpha 1 |
| chr4_-_186733363 | 0.44 |
ENST00000393523.2
ENST00000393528.3 ENST00000449407.2 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr16_-_28374829 | 0.43 |
ENST00000532254.1
|
NPIPB6
|
nuclear pore complex interacting protein family, member B6 |
| chrX_-_63005405 | 0.43 |
ENST00000374878.1
ENST00000437457.2 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr13_+_113633620 | 0.43 |
ENST00000421756.1
ENST00000375601.3 |
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr16_+_83932684 | 0.42 |
ENST00000262430.4
|
MLYCD
|
malonyl-CoA decarboxylase |
| chr6_-_41888843 | 0.42 |
ENST00000434077.1
ENST00000409312.1 |
MED20
|
mediator complex subunit 20 |
| chr17_-_34502410 | 0.42 |
ENST00000398801.3
|
TBC1D3B
|
TBC1 domain family, member 3B |
| chr9_+_90112117 | 0.42 |
ENST00000358077.5
|
DAPK1
|
death-associated protein kinase 1 |
| chr14_-_106539557 | 0.42 |
ENST00000390599.2
|
IGHV1-8
|
immunoglobulin heavy variable 1-8 |
| chr19_-_51920835 | 0.42 |
ENST00000442846.3
ENST00000530476.1 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr3_-_112356944 | 0.42 |
ENST00000461431.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chr12_+_48357401 | 0.42 |
ENST00000429772.2
ENST00000449758.2 |
TMEM106C
|
transmembrane protein 106C |
| chr16_+_14165160 | 0.41 |
ENST00000574998.1
ENST00000571589.1 ENST00000574045.1 |
MKL2
|
MKL/myocardin-like 2 |
| chr1_+_66999268 | 0.41 |
ENST00000371039.1
ENST00000424320.1 |
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr2_+_177053307 | 0.41 |
ENST00000331462.4
|
HOXD1
|
homeobox D1 |
| chr17_-_8534031 | 0.41 |
ENST00000411957.1
ENST00000396239.1 ENST00000379980.4 |
MYH10
|
myosin, heavy chain 10, non-muscle |
| chr19_-_11308190 | 0.40 |
ENST00000586659.1
ENST00000592903.1 ENST00000589359.1 ENST00000588724.1 ENST00000432929.2 |
KANK2
|
KN motif and ankyrin repeat domains 2 |
| chr7_-_8302298 | 0.40 |
ENST00000446305.1
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chrX_-_107019181 | 0.40 |
ENST00000315660.4
ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr9_+_17579084 | 0.40 |
ENST00000380607.4
|
SH3GL2
|
SH3-domain GRB2-like 2 |
| chr17_-_8534067 | 0.40 |
ENST00000360416.3
ENST00000269243.4 |
MYH10
|
myosin, heavy chain 10, non-muscle |
| chr2_-_106054952 | 0.39 |
ENST00000336660.5
ENST00000393352.3 ENST00000607522.1 |
FHL2
|
four and a half LIM domains 2 |
| chr3_-_197282821 | 0.39 |
ENST00000445160.2
ENST00000446746.1 ENST00000432819.1 ENST00000392379.1 ENST00000441275.1 ENST00000392378.2 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr7_-_21985656 | 0.39 |
ENST00000406877.3
|
CDCA7L
|
cell division cycle associated 7-like |
| chr11_-_1629693 | 0.39 |
ENST00000399685.1
|
KRTAP5-3
|
keratin associated protein 5-3 |
| chr4_-_21950356 | 0.38 |
ENST00000447367.2
ENST00000382152.2 |
KCNIP4
|
Kv channel interacting protein 4 |
| chr14_-_106471723 | 0.38 |
ENST00000390595.2
|
IGHV1-3
|
immunoglobulin heavy variable 1-3 |
| chr10_-_49459800 | 0.38 |
ENST00000305531.3
|
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr13_-_52027134 | 0.38 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr15_-_22473353 | 0.38 |
ENST00000557788.2
|
IGHV4OR15-8
|
immunoglobulin heavy variable 4/OR15-8 (non-functional) |
| chr4_-_2420357 | 0.38 |
ENST00000511071.1
ENST00000509171.1 ENST00000290974.2 |
ZFYVE28
|
zinc finger, FYVE domain containing 28 |
| chr19_-_51920952 | 0.38 |
ENST00000356298.5
ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr7_-_120497178 | 0.38 |
ENST00000441017.1
ENST00000424710.1 ENST00000433758.1 |
TSPAN12
|
tetraspanin 12 |
| chr6_-_32077078 | 0.37 |
ENST00000479795.1
|
TNXB
|
tenascin XB |
| chr1_+_78354297 | 0.37 |
ENST00000334785.7
|
NEXN
|
nexilin (F actin binding protein) |
| chr22_-_51016433 | 0.37 |
ENST00000405237.3
|
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr19_-_46272106 | 0.37 |
ENST00000560168.1
|
SIX5
|
SIX homeobox 5 |
| chr4_+_166248775 | 0.37 |
ENST00000261507.6
ENST00000507013.1 ENST00000393766.2 ENST00000504317.1 |
MSMO1
|
methylsterol monooxygenase 1 |
| chr15_+_27112948 | 0.36 |
ENST00000555060.1
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr3_-_134093275 | 0.36 |
ENST00000513145.1
ENST00000422605.2 |
AMOTL2
|
angiomotin like 2 |
| chr9_-_131940526 | 0.36 |
ENST00000372491.2
|
IER5L
|
immediate early response 5-like |
| chr15_-_31283618 | 0.36 |
ENST00000563714.1
|
MTMR10
|
myotubularin related protein 10 |
| chr12_+_48357340 | 0.36 |
ENST00000256686.6
ENST00000549288.1 ENST00000552561.1 ENST00000546749.1 ENST00000552546.1 ENST00000550552.1 |
TMEM106C
|
transmembrane protein 106C |
| chr20_+_48599506 | 0.36 |
ENST00000244050.2
|
SNAI1
|
snail family zinc finger 1 |
| chr11_-_124632179 | 0.35 |
ENST00000278927.5
ENST00000442070.2 ENST00000444566.1 ENST00000435477.1 |
ESAM
|
endothelial cell adhesion molecule |
| chr6_+_159290917 | 0.35 |
ENST00000367072.1
|
C6orf99
|
chromosome 6 open reading frame 99 |
| chr19_-_51920873 | 0.35 |
ENST00000441969.3
ENST00000525998.1 ENST00000436984.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chrX_+_56259316 | 0.35 |
ENST00000468660.1
|
KLF8
|
Kruppel-like factor 8 |
| chr15_+_83776324 | 0.35 |
ENST00000379390.6
ENST00000379386.4 ENST00000565774.1 ENST00000565982.1 |
TM6SF1
|
transmembrane 6 superfamily member 1 |
| chr4_+_40198527 | 0.35 |
ENST00000381799.5
|
RHOH
|
ras homolog family member H |
| chr16_-_21868978 | 0.35 |
ENST00000357370.5
ENST00000451409.1 ENST00000341400.7 ENST00000518761.4 |
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr16_-_29415350 | 0.35 |
ENST00000524087.1
|
NPIPB11
|
nuclear pore complex interacting protein family, member B11 |
| chr15_+_42131011 | 0.35 |
ENST00000458483.1
|
PLA2G4B
|
phospholipase A2, group IVB (cytosolic) |
| chr12_-_49488573 | 0.34 |
ENST00000266991.2
|
DHH
|
desert hedgehog |
| chr17_+_37894179 | 0.34 |
ENST00000577695.1
ENST00000309156.4 ENST00000309185.3 |
GRB7
|
growth factor receptor-bound protein 7 |
| chr10_-_21463116 | 0.34 |
ENST00000417816.2
|
NEBL
|
nebulette |
| chr10_-_75457554 | 0.34 |
ENST00000374094.4
ENST00000443782.2 |
AGAP5
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 5 |
| chr16_-_21436459 | 0.34 |
ENST00000448012.2
ENST00000504841.2 ENST00000419180.2 |
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr17_-_27503770 | 0.34 |
ENST00000533112.1
|
MYO18A
|
myosin XVIIIA |
| chr1_-_59043166 | 0.34 |
ENST00000371225.2
|
TACSTD2
|
tumor-associated calcium signal transducer 2 |
| chr4_+_88928777 | 0.33 |
ENST00000237596.2
|
PKD2
|
polycystic kidney disease 2 (autosomal dominant) |
| chr8_+_98881268 | 0.33 |
ENST00000254898.5
ENST00000524308.1 ENST00000522025.2 |
MATN2
|
matrilin 2 |
| chr7_+_44663908 | 0.33 |
ENST00000543843.1
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr3_-_49851313 | 0.33 |
ENST00000333486.3
|
UBA7
|
ubiquitin-like modifier activating enzyme 7 |
| chr17_+_37793318 | 0.32 |
ENST00000336308.5
|
STARD3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr14_-_74416829 | 0.32 |
ENST00000534936.1
|
FAM161B
|
family with sequence similarity 161, member B |
| chr15_-_20170354 | 0.32 |
ENST00000338912.5
|
IGHV1OR15-9
|
immunoglobulin heavy variable 1/OR15-9 (non-functional) |
| chrX_-_34150428 | 0.32 |
ENST00000346193.3
|
FAM47A
|
family with sequence similarity 47, member A |
| chr4_+_75311019 | 0.32 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chr12_+_56661461 | 0.32 |
ENST00000546544.1
ENST00000553234.1 |
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae) |
| chr17_-_39334460 | 0.32 |
ENST00000377726.2
|
KRTAP4-2
|
keratin associated protein 4-2 |
| chr2_-_50574856 | 0.32 |
ENST00000342183.5
|
NRXN1
|
neurexin 1 |
| chr12_+_125549973 | 0.31 |
ENST00000536752.1
ENST00000261686.6 |
AACS
|
acetoacetyl-CoA synthetase |
| chr8_-_144655141 | 0.31 |
ENST00000398882.3
|
MROH6
|
maestro heat-like repeat family member 6 |
| chr16_+_6533729 | 0.31 |
ENST00000551752.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr14_+_77228532 | 0.31 |
ENST00000167106.4
ENST00000554237.1 |
VASH1
|
vasohibin 1 |
| chr16_+_28763108 | 0.31 |
ENST00000357796.3
ENST00000550983.1 |
NPIPB9
|
nuclear pore complex interacting protein family, member B9 |
| chr17_-_8198636 | 0.31 |
ENST00000577745.1
ENST00000579192.1 ENST00000396278.1 |
SLC25A35
|
solute carrier family 25, member 35 |
| chr9_+_138235095 | 0.31 |
ENST00000320778.2
|
C9orf62
|
chromosome 9 open reading frame 62 |
| chr5_-_19988339 | 0.31 |
ENST00000382275.1
|
CDH18
|
cadherin 18, type 2 |
| chr9_-_127905736 | 0.31 |
ENST00000336505.6
ENST00000373549.4 |
SCAI
|
suppressor of cancer cell invasion |
| chr16_-_21868739 | 0.31 |
ENST00000415645.2
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr12_-_68726052 | 0.31 |
ENST00000540418.1
ENST00000411698.2 ENST00000393543.3 ENST00000303145.7 |
MDM1
|
Mdm1 nuclear protein homolog (mouse) |
| chr3_-_111314230 | 0.31 |
ENST00000317012.4
|
ZBED2
|
zinc finger, BED-type containing 2 |
| chr9_-_116061145 | 0.31 |
ENST00000416588.2
|
RNF183
|
ring finger protein 183 |
| chr20_-_50808525 | 0.31 |
ENST00000216923.4
|
ZFP64
|
ZFP64 zinc finger protein |
| chr8_+_133879193 | 0.31 |
ENST00000377869.1
ENST00000220616.4 |
TG
|
thyroglobulin |
| chr19_-_53770972 | 0.31 |
ENST00000311170.4
|
VN1R4
|
vomeronasal 1 receptor 4 |
| chr3_+_127634312 | 0.31 |
ENST00000407609.3
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr4_+_42399856 | 0.31 |
ENST00000319234.4
|
SHISA3
|
shisa family member 3 |
| chr2_+_33701286 | 0.30 |
ENST00000403687.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr2_+_90198535 | 0.30 |
ENST00000390276.2
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr19_+_42381173 | 0.30 |
ENST00000221972.3
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr15_+_83776137 | 0.30 |
ENST00000322019.9
|
TM6SF1
|
transmembrane 6 superfamily member 1 |
| chr1_+_154377669 | 0.30 |
ENST00000368485.3
ENST00000344086.4 |
IL6R
|
interleukin 6 receptor |
| chr14_+_23842018 | 0.30 |
ENST00000397242.2
ENST00000329715.2 |
IL25
|
interleukin 25 |
| chr1_+_162760513 | 0.30 |
ENST00000367915.1
ENST00000367917.3 ENST00000254521.3 ENST00000367913.1 |
HSD17B7
|
hydroxysteroid (17-beta) dehydrogenase 7 |
| chr14_-_77495007 | 0.30 |
ENST00000238647.3
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like |
| chr5_-_172662303 | 0.30 |
ENST00000517440.1
ENST00000329198.4 |
NKX2-5
|
NK2 homeobox 5 |
| chr8_-_80993010 | 0.30 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr17_+_37793378 | 0.29 |
ENST00000544210.2
ENST00000581894.1 ENST00000394250.4 ENST00000579479.1 ENST00000577248.1 ENST00000580611.1 |
STARD3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr1_+_6845578 | 0.29 |
ENST00000467404.2
ENST00000439411.2 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr2_+_173600514 | 0.29 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr21_+_45770009 | 0.29 |
ENST00000300482.5
ENST00000431901.1 |
TRPM2
|
transient receptor potential cation channel, subfamily M, member 2 |
| chr6_+_35995488 | 0.29 |
ENST00000229795.3
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chrX_+_45364633 | 0.29 |
ENST00000435394.1
ENST00000609127.1 |
RP11-245M24.1
|
RP11-245M24.1 |
| chr1_+_2160134 | 0.29 |
ENST00000378536.4
|
SKI
|
v-ski avian sarcoma viral oncogene homolog |
| chr9_+_90112767 | 0.29 |
ENST00000408954.3
|
DAPK1
|
death-associated protein kinase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MESP1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.6 | GO:0052551 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.5 | 2.1 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.5 | 1.4 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.4 | 2.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.4 | 1.2 | GO:0021571 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.4 | 1.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.3 | 2.5 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.3 | 1.4 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.3 | 1.4 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.3 | 1.0 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.2 | 0.8 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.2 | 0.2 | GO:0097017 | renal protein absorption(GO:0097017) renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) |
| 0.2 | 0.5 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.2 | 0.8 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.2 | 0.5 | GO:0072237 | carbon dioxide transmembrane transport(GO:0035378) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) metanephric proximal tubule development(GO:0072237) |
| 0.2 | 1.0 | GO:0061441 | renal artery morphogenesis(GO:0061441) |
| 0.1 | 1.8 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.1 | 0.7 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.7 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.4 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 | 1.7 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.5 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.4 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 0.5 | GO:0018032 | protein amidation(GO:0018032) |
| 0.1 | 0.3 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.6 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.1 | 1.0 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 1.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 0.3 | GO:0086100 | posterior midgut development(GO:0007497) endothelin receptor signaling pathway(GO:0086100) |
| 0.1 | 0.3 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 0.3 | GO:1990764 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.1 | 0.4 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.4 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.3 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.1 | 0.6 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.5 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.5 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.1 | 0.4 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.1 | 0.3 | GO:0021913 | glandular epithelial cell maturation(GO:0002071) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) type B pancreatic cell maturation(GO:0072560) |
| 0.1 | 0.4 | GO:0036115 | fatty-acyl-CoA catabolic process(GO:0036115) malonyl-CoA metabolic process(GO:2001293) |
| 0.1 | 0.3 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.1 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 0.2 | GO:0001172 | transcription, RNA-templated(GO:0001172) negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.4 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.2 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.2 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.1 | 0.4 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 0.3 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.2 | GO:1905166 | negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.1 | 0.3 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.1 | 0.9 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 0.2 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.1 | 0.6 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.1 | 0.2 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.1 | 0.1 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.1 | 0.2 | GO:0050894 | determination of affect(GO:0050894) |
| 0.1 | 2.0 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.1 | 0.1 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 0.1 | 0.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.3 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.1 | 0.1 | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.1 | 0.2 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.1 | 0.8 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.1 | 0.2 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.2 | GO:0001079 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.1 | 0.4 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 0.3 | GO:0072248 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.1 | 0.6 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.1 | 0.2 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.4 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.1 | 0.3 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.1 | 0.5 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.6 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.1 | 0.2 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.1 | 0.3 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 0.2 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.1 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.4 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.3 | GO:0009439 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.1 | 0.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.1 | 0.5 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 3.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.2 | GO:0060018 | astrocyte fate commitment(GO:0060018) renal vesicle induction(GO:0072034) |
| 0.1 | 0.6 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 9.9 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.3 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.2 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.2 | GO:2000721 | pilomotor reflex(GO:0097195) positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.1 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.2 | GO:0007418 | ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.4 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.9 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 0.2 | GO:0035811 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) negative regulation of urine volume(GO:0035811) |
| 0.0 | 0.4 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.1 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.0 | 0.3 | GO:0009624 | response to nematode(GO:0009624) eosinophil differentiation(GO:0030222) |
| 0.0 | 0.2 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.2 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.1 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.0 | 0.4 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.3 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.2 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.2 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.7 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.2 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.2 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.1 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.0 | 0.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.1 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.0 | 0.5 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.2 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) |
| 0.0 | 1.4 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.7 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.7 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.5 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.3 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0002525 | acute inflammatory response to non-antigenic stimulus(GO:0002525) regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 0.0 | 0.2 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.0 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0070904 | L-ascorbic acid transport(GO:0015882) molecular hydrogen transport(GO:0015993) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.3 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.4 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 0.2 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.2 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.6 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.6 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0015911 | plasma membrane long-chain fatty acid transport(GO:0015911) |
| 0.0 | 0.1 | GO:1903936 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.2 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.1 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.0 | 0.1 | GO:0090301 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.2 | GO:0032445 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.3 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 1.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 | 0.5 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.3 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.0 | 0.6 | GO:0046596 | regulation of viral entry into host cell(GO:0046596) |
| 0.0 | 0.1 | GO:0042853 | L-alanine metabolic process(GO:0042851) L-alanine catabolic process(GO:0042853) |
| 0.0 | 0.2 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) |
| 0.0 | 0.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.1 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.0 | 0.1 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) negative regulation of interleukin-5 production(GO:0032714) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 0.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.2 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.1 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.3 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.6 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.9 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.2 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.4 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.1 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 0.0 | 0.1 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.7 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.4 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.2 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.6 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.2 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.0 | 0.3 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.1 | GO:0032914 | rostrocaudal neural tube patterning(GO:0021903) positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 1.1 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.1 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.0 | 0.0 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.1 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.0 | GO:1904387 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.0 | 0.5 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.1 | GO:0035625 | negative regulation of epinephrine secretion(GO:0032811) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.0 | 1.1 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0031179 | peptide amidation(GO:0001519) peptide modification(GO:0031179) |
| 0.0 | 0.2 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.0 | 0.1 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 1.3 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.3 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.1 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.3 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.2 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.0 | GO:2000672 | regulation of motor neuron apoptotic process(GO:2000671) negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.1 | GO:0048672 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.2 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.3 | 1.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 0.7 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.2 | 0.6 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.2 | 0.9 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.2 | 1.6 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.2 | 4.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.6 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 10.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.8 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 3.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.2 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.1 | 1.0 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.2 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.1 | 1.0 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.2 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.1 | 0.3 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.1 | 0.5 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.2 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.1 | 0.4 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 0.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.6 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 1.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.2 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.4 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 1.5 | GO:0019814 | immunoglobulin complex(GO:0019814) immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.6 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.0 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 1.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.4 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.0 | 0.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 1.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.3 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.2 | 4.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 0.5 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.1 | 0.4 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 0.4 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 1.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.6 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.1 | 0.5 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 1.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.3 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 0.3 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.1 | 0.3 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.3 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 0.7 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.3 | GO:0043398 | HLH domain binding(GO:0043398) calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 0.3 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.1 | 0.7 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.1 | 0.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.8 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.2 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.1 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.3 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.5 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 1.6 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 0.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.2 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.1 | 0.7 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 0.2 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.1 | 0.2 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.1 | 1.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.4 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.1 | 0.2 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.1 | 0.6 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.1 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 3.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.3 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.5 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.0 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.2 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.2 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.3 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.2 | GO:0030395 | lactose binding(GO:0030395) |
| 0.0 | 2.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.2 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.2 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.6 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.2 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.2 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.3 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.3 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 1.0 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.3 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.1 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.4 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0004368 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 1.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.0 | 0.2 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.6 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.3 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.2 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.3 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.2 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.5 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 1.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.9 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.1 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.0 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.2 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.1 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.0 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 1.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 4.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 1.1 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 1.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.9 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.0 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.3 | PID NOTCH PATHWAY | Notch signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 3.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 0.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 3.0 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.7 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.0 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 2.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.0 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.1 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |