Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for MSX1
Z-value: 0.47
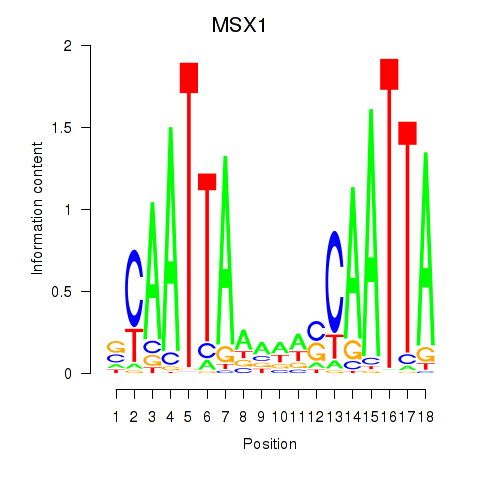
Transcription factors associated with MSX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MSX1
|
ENSG00000163132.6 | msh homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MSX1 | hg19_v2_chr4_+_4861385_4861398 | -0.01 | 9.4e-01 | Click! |
Activity profile of MSX1 motif
Sorted Z-values of MSX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_43619796 | 1.06 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr10_-_101380121 | 0.89 |
ENST00000370495.4
|
SLC25A28
|
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr6_+_28249299 | 0.85 |
ENST00000405948.2
|
PGBD1
|
piggyBac transposable element derived 1 |
| chr6_+_28249332 | 0.85 |
ENST00000259883.3
|
PGBD1
|
piggyBac transposable element derived 1 |
| chr5_+_121647386 | 0.73 |
ENST00000542191.1
ENST00000506272.1 ENST00000508681.1 ENST00000509154.2 |
SNCAIP
|
synuclein, alpha interacting protein |
| chr22_+_41697520 | 0.58 |
ENST00000352645.4
|
ZC3H7B
|
zinc finger CCCH-type containing 7B |
| chr12_-_52911718 | 0.57 |
ENST00000548409.1
|
KRT5
|
keratin 5 |
| chr19_+_3762645 | 0.53 |
ENST00000330133.4
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr17_-_37934466 | 0.53 |
ENST00000583368.1
|
IKZF3
|
IKAROS family zinc finger 3 (Aiolos) |
| chr9_+_115913222 | 0.52 |
ENST00000259392.3
|
SLC31A2
|
solute carrier family 31 (copper transporter), member 2 |
| chr2_-_145278475 | 0.49 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr5_-_11588907 | 0.46 |
ENST00000513598.1
ENST00000503622.1 |
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr6_+_26240561 | 0.44 |
ENST00000377745.2
|
HIST1H4F
|
histone cluster 1, H4f |
| chr3_-_190040223 | 0.41 |
ENST00000295522.3
|
CLDN1
|
claudin 1 |
| chr3_-_123339343 | 0.36 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr3_-_123339418 | 0.35 |
ENST00000583087.1
|
MYLK
|
myosin light chain kinase |
| chr11_-_5345582 | 0.35 |
ENST00000328813.2
|
OR51B2
|
olfactory receptor, family 51, subfamily B, member 2 |
| chr10_+_13141585 | 0.35 |
ENST00000378764.2
|
OPTN
|
optineurin |
| chr3_-_123512688 | 0.34 |
ENST00000475616.1
|
MYLK
|
myosin light chain kinase |
| chrX_+_1710484 | 0.34 |
ENST00000313871.3
ENST00000381261.3 |
AKAP17A
|
A kinase (PRKA) anchor protein 17A |
| chr1_-_149908217 | 0.33 |
ENST00000369140.3
|
MTMR11
|
myotubularin related protein 11 |
| chr8_+_30244580 | 0.31 |
ENST00000523115.1
ENST00000519647.1 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr18_+_61637159 | 0.30 |
ENST00000397985.2
ENST00000353706.2 ENST00000542677.1 ENST00000397988.3 ENST00000448851.1 |
SERPINB8
|
serpin peptidase inhibitor, clade B (ovalbumin), member 8 |
| chr2_+_196313239 | 0.30 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chr10_+_13142225 | 0.29 |
ENST00000378747.3
|
OPTN
|
optineurin |
| chr19_+_3762703 | 0.29 |
ENST00000589174.1
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr14_-_101295407 | 0.29 |
ENST00000596284.1
|
AL117190.2
|
AL117190.2 |
| chr4_+_146539415 | 0.29 |
ENST00000281317.5
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr6_+_29141311 | 0.28 |
ENST00000377167.2
|
OR2J2
|
olfactory receptor, family 2, subfamily J, member 2 |
| chr11_-_128894053 | 0.27 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr22_+_51176624 | 0.27 |
ENST00000216139.5
ENST00000529621.1 |
ACR
|
acrosin |
| chr17_-_30228678 | 0.26 |
ENST00000261708.4
|
UTP6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr3_+_171561127 | 0.26 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr14_+_77582905 | 0.24 |
ENST00000557408.1
|
TMEM63C
|
transmembrane protein 63C |
| chr7_-_16840820 | 0.24 |
ENST00000450569.1
|
AGR2
|
anterior gradient 2 |
| chr8_-_25281747 | 0.24 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr12_+_9980069 | 0.24 |
ENST00000354855.3
ENST00000324214.4 ENST00000279544.3 |
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr18_-_5396271 | 0.24 |
ENST00000579951.1
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr19_+_16059818 | 0.22 |
ENST00000322107.1
|
OR10H4
|
olfactory receptor, family 10, subfamily H, member 4 |
| chr7_-_20256965 | 0.22 |
ENST00000400331.5
ENST00000332878.4 |
MACC1
|
metastasis associated in colon cancer 1 |
| chr12_+_105380073 | 0.22 |
ENST00000552951.1
ENST00000280749.5 |
C12orf45
|
chromosome 12 open reading frame 45 |
| chr15_-_77712477 | 0.21 |
ENST00000560626.2
|
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr11_-_102496063 | 0.20 |
ENST00000260228.2
|
MMP20
|
matrix metallopeptidase 20 |
| chr1_+_224370873 | 0.20 |
ENST00000323699.4
ENST00000391877.3 |
DEGS1
|
delta(4)-desaturase, sphingolipid 1 |
| chr17_-_8113542 | 0.20 |
ENST00000578549.1
ENST00000535053.1 ENST00000582368.1 |
AURKB
|
aurora kinase B |
| chr15_-_95870310 | 0.20 |
ENST00000508732.2
|
CTD-2536I1.1
|
CTD-2536I1.1 |
| chr11_-_92931098 | 0.19 |
ENST00000326402.4
|
SLC36A4
|
solute carrier family 36 (proton/amino acid symporter), member 4 |
| chr2_-_209028300 | 0.19 |
ENST00000304502.4
|
CRYGA
|
crystallin, gamma A |
| chr7_-_32338917 | 0.19 |
ENST00000396193.1
|
PDE1C
|
phosphodiesterase 1C, calmodulin-dependent 70kDa |
| chr4_-_120243545 | 0.19 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr9_-_128246769 | 0.19 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr5_+_102594403 | 0.18 |
ENST00000319933.2
|
C5orf30
|
chromosome 5 open reading frame 30 |
| chr2_-_88927092 | 0.18 |
ENST00000303236.3
|
EIF2AK3
|
eukaryotic translation initiation factor 2-alpha kinase 3 |
| chr9_+_133539981 | 0.18 |
ENST00000253008.2
|
PRDM12
|
PR domain containing 12 |
| chr12_+_54422142 | 0.17 |
ENST00000243108.4
|
HOXC6
|
homeobox C6 |
| chr19_+_18303992 | 0.17 |
ENST00000599612.2
|
MPV17L2
|
MPV17 mitochondrial membrane protein-like 2 |
| chr2_-_158345462 | 0.16 |
ENST00000439355.1
ENST00000540637.1 |
CYTIP
|
cytohesin 1 interacting protein |
| chr20_+_56964253 | 0.16 |
ENST00000395802.3
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr10_-_29923893 | 0.16 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr14_-_75389975 | 0.16 |
ENST00000555647.1
ENST00000557413.1 |
RPS6KL1
|
ribosomal protein S6 kinase-like 1 |
| chr5_+_67586465 | 0.16 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr15_+_51669444 | 0.15 |
ENST00000396399.2
|
GLDN
|
gliomedin |
| chr3_+_114012819 | 0.15 |
ENST00000383671.3
|
TIGIT
|
T cell immunoreceptor with Ig and ITIM domains |
| chr1_-_160492994 | 0.15 |
ENST00000368055.1
ENST00000368057.3 ENST00000368059.3 |
SLAMF6
|
SLAM family member 6 |
| chr10_-_43892668 | 0.15 |
ENST00000544000.1
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr17_-_39150385 | 0.14 |
ENST00000391586.1
|
KRTAP3-3
|
keratin associated protein 3-3 |
| chr1_+_160160283 | 0.14 |
ENST00000368079.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr3_-_47555167 | 0.13 |
ENST00000296149.4
|
ELP6
|
elongator acetyltransferase complex subunit 6 |
| chr1_+_160160346 | 0.13 |
ENST00000368078.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr9_+_44868935 | 0.13 |
ENST00000448436.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr13_-_38172863 | 0.13 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chr7_+_148287657 | 0.12 |
ENST00000307003.2
|
C7orf33
|
chromosome 7 open reading frame 33 |
| chr17_-_48785216 | 0.12 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr1_+_46805832 | 0.12 |
ENST00000474844.1
|
NSUN4
|
NOP2/Sun domain family, member 4 |
| chr10_+_135204338 | 0.12 |
ENST00000468317.2
|
RP11-108K14.8
|
Mitochondrial GTPase 1 |
| chr10_+_32873190 | 0.12 |
ENST00000375025.4
|
C10orf68
|
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
| chr8_+_97597148 | 0.12 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr12_+_7864021 | 0.12 |
ENST00000345088.2
|
DPPA3
|
developmental pluripotency associated 3 |
| chr6_-_32784687 | 0.12 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr3_-_21792838 | 0.12 |
ENST00000281523.2
|
ZNF385D
|
zinc finger protein 385D |
| chr11_-_124190184 | 0.12 |
ENST00000357438.2
|
OR8D2
|
olfactory receptor, family 8, subfamily D, member 2 |
| chr1_+_158815588 | 0.12 |
ENST00000438394.1
|
MNDA
|
myeloid cell nuclear differentiation antigen |
| chr5_-_147162078 | 0.11 |
ENST00000507386.1
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2 |
| chr6_+_134758827 | 0.11 |
ENST00000431422.1
|
LINC01010
|
long intergenic non-protein coding RNA 1010 |
| chr16_-_9030515 | 0.11 |
ENST00000535863.1
ENST00000381886.4 |
USP7
|
ubiquitin specific peptidase 7 (herpes virus-associated) |
| chr11_+_45825616 | 0.11 |
ENST00000442528.2
ENST00000456334.1 ENST00000526817.1 |
SLC35C1
|
solute carrier family 35 (GDP-fucose transporter), member C1 |
| chr22_+_30477000 | 0.11 |
ENST00000403975.1
|
HORMAD2
|
HORMA domain containing 2 |
| chr6_+_131958436 | 0.10 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr20_+_5987890 | 0.10 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr11_+_61197508 | 0.10 |
ENST00000541135.1
ENST00000301761.2 |
RP11-286N22.8
SDHAF2
|
Uncharacterized protein succinate dehydrogenase complex assembly factor 2 |
| chr2_-_109605663 | 0.10 |
ENST00000409271.1
ENST00000258443.2 ENST00000376651.1 |
EDAR
|
ectodysplasin A receptor |
| chr16_+_69345243 | 0.09 |
ENST00000254950.11
|
VPS4A
|
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
| chr12_+_106751436 | 0.09 |
ENST00000228347.4
|
POLR3B
|
polymerase (RNA) III (DNA directed) polypeptide B |
| chr8_-_121824374 | 0.09 |
ENST00000517992.1
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr12_-_52604607 | 0.09 |
ENST00000551894.1
ENST00000553017.1 |
C12orf80
|
chromosome 12 open reading frame 80 |
| chr18_-_3874271 | 0.09 |
ENST00000400149.3
ENST00000400155.1 ENST00000400150.3 |
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr8_+_117963190 | 0.09 |
ENST00000427715.2
|
SLC30A8
|
solute carrier family 30 (zinc transporter), member 8 |
| chr18_-_70532906 | 0.08 |
ENST00000299430.2
ENST00000397929.1 |
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr4_-_57547454 | 0.08 |
ENST00000556376.2
|
HOPX
|
HOP homeobox |
| chr8_-_72274095 | 0.08 |
ENST00000303824.7
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr6_+_118869452 | 0.08 |
ENST00000357525.5
|
PLN
|
phospholamban |
| chr19_-_53757941 | 0.08 |
ENST00000594517.1
ENST00000601413.1 ENST00000594681.1 |
ZNF677
|
zinc finger protein 677 |
| chr14_+_31046959 | 0.08 |
ENST00000547532.1
ENST00000555429.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr2_+_12246664 | 0.08 |
ENST00000449986.1
|
AC096559.1
|
AC096559.1 |
| chr15_-_76304731 | 0.08 |
ENST00000394907.3
|
NRG4
|
neuregulin 4 |
| chr5_+_82767487 | 0.08 |
ENST00000343200.5
ENST00000342785.4 |
VCAN
|
versican |
| chr1_-_2461684 | 0.07 |
ENST00000378453.3
|
HES5
|
hes family bHLH transcription factor 5 |
| chr18_-_3874247 | 0.07 |
ENST00000581699.1
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr4_+_113568207 | 0.07 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr1_-_226595741 | 0.07 |
ENST00000366794.5
ENST00000366792.1 ENST00000366791.5 |
PARP1
|
poly (ADP-ribose) polymerase 1 |
| chr6_+_26020672 | 0.07 |
ENST00000357647.3
|
HIST1H3A
|
histone cluster 1, H3a |
| chr6_+_28092338 | 0.07 |
ENST00000340487.4
|
ZSCAN16
|
zinc finger and SCAN domain containing 16 |
| chr16_+_21689835 | 0.07 |
ENST00000286149.4
ENST00000388958.3 |
OTOA
|
otoancorin |
| chr1_-_72566613 | 0.07 |
ENST00000306821.3
|
NEGR1
|
neuronal growth regulator 1 |
| chr11_-_85376121 | 0.07 |
ENST00000527447.1
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr20_+_56964169 | 0.07 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr18_-_47721447 | 0.07 |
ENST00000285039.7
|
MYO5B
|
myosin VB |
| chr21_+_33671160 | 0.07 |
ENST00000303645.5
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr2_-_77749474 | 0.07 |
ENST00000409093.1
ENST00000409088.3 |
LRRTM4
|
leucine rich repeat transmembrane neuronal 4 |
| chr5_+_82767583 | 0.06 |
ENST00000512590.2
ENST00000513960.1 ENST00000513984.1 ENST00000502527.2 |
VCAN
|
versican |
| chr10_+_90346519 | 0.06 |
ENST00000371939.3
|
LIPJ
|
lipase, family member J |
| chr5_-_138739739 | 0.06 |
ENST00000514983.1
ENST00000507779.2 ENST00000451821.2 ENST00000450845.2 ENST00000509959.1 ENST00000302091.5 |
SPATA24
|
spermatogenesis associated 24 |
| chr5_+_150404904 | 0.06 |
ENST00000521632.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chrX_-_14891150 | 0.06 |
ENST00000452869.1
ENST00000398334.1 ENST00000324138.3 |
FANCB
|
Fanconi anemia, complementation group B |
| chr2_+_202098203 | 0.06 |
ENST00000450491.1
ENST00000440732.1 ENST00000392258.3 |
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chrY_-_2655644 | 0.05 |
ENST00000525526.2
ENST00000534739.2 ENST00000383070.1 |
SRY
|
sex determining region Y |
| chrX_+_134654540 | 0.05 |
ENST00000370752.4
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr3_+_93781728 | 0.05 |
ENST00000314622.4
|
NSUN3
|
NOP2/Sun domain family, member 3 |
| chr14_+_52327109 | 0.05 |
ENST00000335281.4
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr12_-_82752565 | 0.05 |
ENST00000256151.7
|
CCDC59
|
coiled-coil domain containing 59 |
| chr16_-_71518504 | 0.05 |
ENST00000565100.2
|
ZNF19
|
zinc finger protein 19 |
| chr11_+_61197572 | 0.05 |
ENST00000542074.1
ENST00000534878.1 ENST00000537782.1 ENST00000543265.1 |
SDHAF2
|
succinate dehydrogenase complex assembly factor 2 |
| chr2_+_158114051 | 0.05 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr11_+_62037622 | 0.05 |
ENST00000227918.2
ENST00000525380.1 |
SCGB2A2
|
secretoglobin, family 2A, member 2 |
| chr13_-_88323218 | 0.05 |
ENST00000436290.2
ENST00000453832.2 ENST00000606590.1 |
MIR4500HG
|
MIR4500 host gene (non-protein coding) |
| chr12_-_11150474 | 0.05 |
ENST00000538986.1
|
TAS2R20
|
taste receptor, type 2, member 20 |
| chr19_-_7812446 | 0.05 |
ENST00000394173.4
ENST00000301357.8 |
CD209
|
CD209 molecule |
| chr14_+_55595762 | 0.05 |
ENST00000254301.9
|
LGALS3
|
lectin, galactoside-binding, soluble, 3 |
| chr19_-_53662257 | 0.05 |
ENST00000599096.1
ENST00000334197.7 ENST00000597183.1 ENST00000601804.1 ENST00000601469.2 ENST00000452676.2 |
ZNF347
|
zinc finger protein 347 |
| chr2_-_31361543 | 0.04 |
ENST00000349752.5
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr2_-_169746878 | 0.04 |
ENST00000282074.2
|
SPC25
|
SPC25, NDC80 kinetochore complex component |
| chr11_+_12766583 | 0.04 |
ENST00000361985.2
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr10_-_32667660 | 0.04 |
ENST00000375110.2
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila) |
| chr14_+_50291993 | 0.04 |
ENST00000595378.1
|
AL627171.2
|
HCG1786899; PRO2610; Uncharacterized protein |
| chr6_-_109777128 | 0.04 |
ENST00000358807.3
ENST00000358577.3 |
MICAL1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr19_-_39805976 | 0.03 |
ENST00000248668.4
|
LRFN1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr14_-_75389925 | 0.03 |
ENST00000556776.1
|
RPS6KL1
|
ribosomal protein S6 kinase-like 1 |
| chr4_+_119810134 | 0.03 |
ENST00000434046.2
|
SYNPO2
|
synaptopodin 2 |
| chr6_-_32557610 | 0.03 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr9_+_130026756 | 0.03 |
ENST00000314904.5
ENST00000373387.4 |
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr4_+_5527117 | 0.03 |
ENST00000505296.1
|
C4orf6
|
chromosome 4 open reading frame 6 |
| chr4_+_5526883 | 0.03 |
ENST00000195455.2
|
C4orf6
|
chromosome 4 open reading frame 6 |
| chr11_+_60681346 | 0.03 |
ENST00000227525.3
|
TMEM109
|
transmembrane protein 109 |
| chr6_+_6588316 | 0.03 |
ENST00000379953.2
|
LY86
|
lymphocyte antigen 86 |
| chr7_+_138818490 | 0.03 |
ENST00000430935.1
ENST00000495038.1 ENST00000474035.2 ENST00000478836.2 ENST00000464848.1 ENST00000343187.4 |
TTC26
|
tetratricopeptide repeat domain 26 |
| chr4_-_164534657 | 0.02 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr1_+_8021954 | 0.02 |
ENST00000377491.1
ENST00000377488.1 |
PARK7
|
parkinson protein 7 |
| chr21_+_17792672 | 0.02 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr14_+_52327350 | 0.02 |
ENST00000555472.1
ENST00000556766.1 |
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr11_-_114430570 | 0.02 |
ENST00000251921.2
|
NXPE1
|
neurexophilin and PC-esterase domain family, member 1 |
| chr10_+_135050908 | 0.02 |
ENST00000325980.9
|
VENTX
|
VENT homeobox |
| chr6_+_131894284 | 0.02 |
ENST00000368087.3
ENST00000356962.2 |
ARG1
|
arginase 1 |
| chr5_+_82767284 | 0.02 |
ENST00000265077.3
|
VCAN
|
versican |
| chr4_-_100140331 | 0.02 |
ENST00000407820.2
ENST00000394897.1 ENST00000508558.1 ENST00000394899.2 |
ADH6
|
alcohol dehydrogenase 6 (class V) |
| chr1_-_19426149 | 0.02 |
ENST00000429347.2
|
UBR4
|
ubiquitin protein ligase E3 component n-recognin 4 |
| chr7_+_2636522 | 0.02 |
ENST00000423196.1
|
IQCE
|
IQ motif containing E |
| chr19_+_55043977 | 0.02 |
ENST00000335056.3
|
KIR3DX1
|
killer cell immunoglobulin-like receptor, three domains, X1 |
| chr20_+_52824367 | 0.02 |
ENST00000371419.2
|
PFDN4
|
prefoldin subunit 4 |
| chr20_+_5731083 | 0.02 |
ENST00000445603.1
ENST00000442185.1 |
C20orf196
|
chromosome 20 open reading frame 196 |
| chr1_-_109935819 | 0.01 |
ENST00000538502.1
|
SORT1
|
sortilin 1 |
| chr14_+_55595960 | 0.01 |
ENST00000554715.1
|
LGALS3
|
lectin, galactoside-binding, soluble, 3 |
| chr12_-_10541575 | 0.01 |
ENST00000540818.1
|
KLRK1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr19_+_29456034 | 0.01 |
ENST00000589921.1
|
LINC00906
|
long intergenic non-protein coding RNA 906 |
| chr2_+_168675182 | 0.01 |
ENST00000305861.1
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr5_+_140762268 | 0.01 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr4_+_159443024 | 0.01 |
ENST00000448688.2
|
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chr19_-_53758094 | 0.01 |
ENST00000601828.1
ENST00000598513.1 ENST00000599012.1 ENST00000333952.4 ENST00000598806.1 |
ZNF677
|
zinc finger protein 677 |
| chr4_+_119809984 | 0.00 |
ENST00000307142.4
ENST00000448416.2 ENST00000429713.2 |
SYNPO2
|
synaptopodin 2 |
| chr9_-_86593238 | 0.00 |
ENST00000351839.3
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K |
| chr19_-_7812397 | 0.00 |
ENST00000593660.1
ENST00000354397.6 ENST00000593821.1 ENST00000602261.1 ENST00000315591.8 ENST00000394161.5 ENST00000204801.8 ENST00000601256.1 ENST00000601951.1 ENST00000315599.7 |
CD209
|
CD209 molecule |
| chr13_-_81801115 | 0.00 |
ENST00000567258.1
|
LINC00564
|
long intergenic non-protein coding RNA 564 |
| chr1_-_54405773 | 0.00 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr17_-_42441204 | 0.00 |
ENST00000293443.7
|
FAM171A2
|
family with sequence similarity 171, member A2 |
| chr1_+_74701062 | 0.00 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
Network of associatons between targets according to the STRING database.
First level regulatory network of MSX1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.3 | 1.1 | GO:0055099 | detection of endogenous stimulus(GO:0009726) response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 0.4 | GO:1903348 | cellular response to lead ion(GO:0071284) positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.1 | 1.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.5 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.3 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.6 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.2 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.1 | 0.2 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 0.2 | GO:1990737 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.1 | 0.2 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.1 | 0.5 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.3 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.1 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.3 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.2 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.2 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:1901536 | negative regulation of DNA demethylation(GO:1901536) regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.2 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.1 | GO:1902081 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.0 | 0.1 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) bone regeneration(GO:1990523) |
| 0.0 | 0.1 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.3 | GO:0014894 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0036446 | peptidyl-serine ADP-ribosylation(GO:0018312) myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 | 0.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.7 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.3 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.3 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0052148 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.1 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.5 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.1 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 0.2 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) negative regulation of immunological synapse formation(GO:2000521) |
| 0.0 | 0.1 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 1.2 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 1.1 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 2.2 | GO:0055037 | recycling endosome(GO:0055037) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 1.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.9 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 0.3 | GO:0004040 | amidase activity(GO:0004040) |
| 0.1 | 0.2 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.1 | 0.5 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.3 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0043337 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.1 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.1 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.1 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 1.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.0 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.2 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |