Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for MYBL1
Z-value: 0.54
Transcription factors associated with MYBL1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYBL1
|
ENSG00000185697.12 | MYB proto-oncogene like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYBL1 | hg19_v2_chr8_-_67525473_67525518 | -0.13 | 5.4e-01 | Click! |
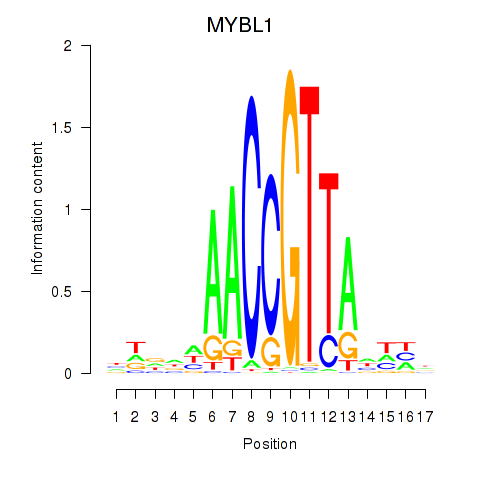
Activity profile of MYBL1 motif
Sorted Z-values of MYBL1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_32455111 | 0.68 |
ENST00000543737.1
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr6_+_26240561 | 0.64 |
ENST00000377745.2
|
HIST1H4F
|
histone cluster 1, H4f |
| chr2_-_133427767 | 0.63 |
ENST00000397463.2
|
LYPD1
|
LY6/PLAUR domain containing 1 |
| chr11_+_20409070 | 0.57 |
ENST00000331079.6
|
PRMT3
|
protein arginine methyltransferase 3 |
| chr3_-_148804275 | 0.57 |
ENST00000392912.2
ENST00000465259.1 ENST00000310053.5 ENST00000494055.1 |
HLTF
|
helicase-like transcription factor |
| chr3_+_58223228 | 0.54 |
ENST00000478253.1
ENST00000295962.4 |
ABHD6
|
abhydrolase domain containing 6 |
| chr11_-_27723158 | 0.54 |
ENST00000395980.2
|
BDNF
|
brain-derived neurotrophic factor |
| chr2_-_190627481 | 0.53 |
ENST00000264151.5
ENST00000520350.1 ENST00000521630.1 ENST00000517895.1 |
OSGEPL1
|
O-sialoglycoprotein endopeptidase-like 1 |
| chr5_-_88179302 | 0.52 |
ENST00000504921.2
|
MEF2C
|
myocyte enhancer factor 2C |
| chr6_-_29324054 | 0.51 |
ENST00000543825.1
|
OR5V1
|
olfactory receptor, family 5, subfamily V, member 1 |
| chr8_+_124084899 | 0.48 |
ENST00000287380.1
ENST00000309336.3 ENST00000519418.1 ENST00000327098.5 ENST00000522420.1 ENST00000521676.1 ENST00000378080.2 |
TBC1D31
|
TBC1 domain family, member 31 |
| chr11_+_18433840 | 0.46 |
ENST00000541669.1
ENST00000280704.4 |
LDHC
|
lactate dehydrogenase C |
| chr13_+_114567131 | 0.46 |
ENST00000608651.1
|
GAS6-AS2
|
GAS6 antisense RNA 2 (head to head) |
| chr13_+_37393351 | 0.46 |
ENST00000255476.2
|
RFXAP
|
regulatory factor X-associated protein |
| chr1_+_163291680 | 0.44 |
ENST00000450453.2
ENST00000524800.1 ENST00000442820.1 ENST00000367900.3 |
NUF2
|
NUF2, NDC80 kinetochore complex component |
| chr14_-_105420241 | 0.43 |
ENST00000557457.1
|
AHNAK2
|
AHNAK nucleoprotein 2 |
| chr1_+_63989004 | 0.42 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr5_-_88179017 | 0.42 |
ENST00000514028.1
ENST00000514015.1 ENST00000503075.1 ENST00000437473.2 |
MEF2C
|
myocyte enhancer factor 2C |
| chr20_+_34802295 | 0.42 |
ENST00000432603.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr1_+_210406121 | 0.42 |
ENST00000367012.3
|
SERTAD4
|
SERTA domain containing 4 |
| chr17_-_30668887 | 0.42 |
ENST00000581747.1
ENST00000583334.1 ENST00000580558.1 |
C17orf75
|
chromosome 17 open reading frame 75 |
| chr6_+_89855765 | 0.41 |
ENST00000275072.4
|
PM20D2
|
peptidase M20 domain containing 2 |
| chr12_-_14996355 | 0.40 |
ENST00000228936.4
|
ART4
|
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr8_+_95565947 | 0.40 |
ENST00000523011.1
|
RP11-267M23.4
|
RP11-267M23.4 |
| chr1_-_151882031 | 0.40 |
ENST00000489410.1
|
THEM4
|
thioesterase superfamily member 4 |
| chr8_-_81083341 | 0.38 |
ENST00000519303.2
|
TPD52
|
tumor protein D52 |
| chr7_+_12726474 | 0.36 |
ENST00000396662.1
ENST00000356797.3 ENST00000396664.2 |
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr17_-_30669138 | 0.36 |
ENST00000225805.4
ENST00000577809.1 |
C17orf75
|
chromosome 17 open reading frame 75 |
| chr9_+_104296122 | 0.36 |
ENST00000389120.3
|
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr2_+_69969106 | 0.35 |
ENST00000409920.1
ENST00000394295.4 ENST00000536030.1 |
ANXA4
|
annexin A4 |
| chr16_-_20753114 | 0.35 |
ENST00000396083.2
|
THUMPD1
|
THUMP domain containing 1 |
| chr12_-_123201337 | 0.34 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr5_+_61708582 | 0.33 |
ENST00000325324.6
|
IPO11
|
importin 11 |
| chr9_+_35906176 | 0.32 |
ENST00000354323.2
|
HRCT1
|
histidine rich carboxyl terminus 1 |
| chr3_+_173116225 | 0.32 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr10_+_5135981 | 0.30 |
ENST00000380554.3
|
AKR1C3
|
aldo-keto reductase family 1, member C3 |
| chr13_-_33760216 | 0.30 |
ENST00000255486.4
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr8_-_81083731 | 0.30 |
ENST00000379096.5
|
TPD52
|
tumor protein D52 |
| chr19_+_52839490 | 0.30 |
ENST00000321287.8
|
ZNF610
|
zinc finger protein 610 |
| chrX_-_104465358 | 0.29 |
ENST00000372578.3
ENST00000372575.1 ENST00000413579.1 |
TEX13A
|
testis expressed 13A |
| chr19_+_58545434 | 0.29 |
ENST00000282326.1
ENST00000601162.1 |
ZSCAN1
|
zinc finger and SCAN domain containing 1 |
| chr8_+_96037205 | 0.29 |
ENST00000396124.4
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr7_+_64254766 | 0.29 |
ENST00000307355.7
ENST00000359735.3 |
ZNF138
|
zinc finger protein 138 |
| chr7_-_123198284 | 0.28 |
ENST00000355749.2
|
NDUFA5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr12_-_76478446 | 0.28 |
ENST00000393263.3
ENST00000548044.1 ENST00000547704.1 ENST00000431879.3 ENST00000549596.1 ENST00000550934.1 ENST00000551600.1 ENST00000547479.1 ENST00000547773.1 ENST00000544816.1 ENST00000542344.1 ENST00000548273.1 |
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr7_-_123197733 | 0.28 |
ENST00000470123.1
ENST00000471770.1 |
NDUFA5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr6_+_88182643 | 0.28 |
ENST00000369556.3
ENST00000544441.1 ENST00000369552.4 ENST00000369557.5 |
SLC35A1
|
solute carrier family 35 (CMP-sialic acid transporter), member A1 |
| chr17_+_47865917 | 0.28 |
ENST00000259021.4
ENST00000454930.2 ENST00000509773.1 ENST00000510819.1 ENST00000424009.2 |
KAT7
|
K(lysine) acetyltransferase 7 |
| chr5_-_75919253 | 0.28 |
ENST00000296641.4
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr19_+_52839515 | 0.28 |
ENST00000403906.3
ENST00000601151.1 |
ZNF610
|
zinc finger protein 610 |
| chr13_+_32838801 | 0.28 |
ENST00000542859.1
|
FRY
|
furry homolog (Drosophila) |
| chr19_-_52551814 | 0.28 |
ENST00000594154.1
ENST00000598745.1 ENST00000597273.1 |
ZNF432
|
zinc finger protein 432 |
| chr19_-_53360853 | 0.28 |
ENST00000596559.1
ENST00000594602.1 ENST00000595646.1 ENST00000597924.1 ENST00000396409.4 ENST00000390651.4 ENST00000243639.4 |
ZNF28
ZNF468
|
zinc finger protein 28 zinc finger protein 468 |
| chr1_+_163291732 | 0.27 |
ENST00000271452.3
|
NUF2
|
NUF2, NDC80 kinetochore complex component |
| chr12_-_27167233 | 0.27 |
ENST00000535819.1
ENST00000543803.1 ENST00000535423.1 ENST00000539741.1 ENST00000343028.4 ENST00000545600.1 ENST00000543088.1 |
TM7SF3
|
transmembrane 7 superfamily member 3 |
| chr19_+_37096194 | 0.27 |
ENST00000460670.1
ENST00000292928.2 ENST00000439428.1 |
ZNF382
|
zinc finger protein 382 |
| chr15_-_79103757 | 0.26 |
ENST00000388820.4
|
ADAMTS7
|
ADAM metallopeptidase with thrombospondin type 1 motif, 7 |
| chr5_+_156607829 | 0.26 |
ENST00000422843.3
|
ITK
|
IL2-inducible T-cell kinase |
| chr3_-_3221358 | 0.26 |
ENST00000424814.1
ENST00000450014.1 ENST00000231948.4 ENST00000432408.2 |
CRBN
|
cereblon |
| chr11_+_43380459 | 0.26 |
ENST00000299240.6
ENST00000039989.4 |
TTC17
|
tetratricopeptide repeat domain 17 |
| chr4_+_123747834 | 0.25 |
ENST00000264498.3
|
FGF2
|
fibroblast growth factor 2 (basic) |
| chr4_+_103790462 | 0.25 |
ENST00000503643.1
|
CISD2
|
CDGSH iron sulfur domain 2 |
| chr13_-_41768654 | 0.25 |
ENST00000379483.3
|
KBTBD7
|
kelch repeat and BTB (POZ) domain containing 7 |
| chr4_+_79567362 | 0.25 |
ENST00000512322.1
|
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr2_-_47382414 | 0.24 |
ENST00000294947.2
|
C2orf61
|
chromosome 2 open reading frame 61 |
| chr12_+_7053228 | 0.24 |
ENST00000540506.2
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr17_-_42019836 | 0.24 |
ENST00000225992.3
|
PPY
|
pancreatic polypeptide |
| chr7_+_23221438 | 0.24 |
ENST00000258742.5
|
NUPL2
|
nucleoporin like 2 |
| chr3_-_49314640 | 0.24 |
ENST00000436325.1
|
C3orf62
|
chromosome 3 open reading frame 62 |
| chr2_+_88047606 | 0.24 |
ENST00000359481.4
|
PLGLB2
|
plasminogen-like B2 |
| chr3_+_49977440 | 0.24 |
ENST00000442092.1
ENST00000266022.4 ENST00000443081.1 |
RBM6
|
RNA binding motif protein 6 |
| chr7_-_16685422 | 0.24 |
ENST00000306999.2
|
ANKMY2
|
ankyrin repeat and MYND domain containing 2 |
| chr3_-_160117301 | 0.24 |
ENST00000326448.7
ENST00000498409.1 ENST00000475677.1 ENST00000478536.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr5_-_147211226 | 0.24 |
ENST00000296695.5
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1 |
| chr2_+_201936458 | 0.24 |
ENST00000237889.4
|
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr3_+_49977490 | 0.23 |
ENST00000539992.1
|
RBM6
|
RNA binding motif protein 6 |
| chr1_+_97187318 | 0.23 |
ENST00000609116.1
ENST00000370198.1 ENST00000370197.1 ENST00000426398.2 ENST00000394184.3 |
PTBP2
|
polypyrimidine tract binding protein 2 |
| chr8_+_92082424 | 0.23 |
ENST00000285420.4
ENST00000404789.3 |
OTUD6B
|
OTU domain containing 6B |
| chr7_+_7222233 | 0.23 |
ENST00000436587.2
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr10_-_23003460 | 0.23 |
ENST00000376573.4
|
PIP4K2A
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
| chrX_+_105066524 | 0.23 |
ENST00000243300.9
ENST00000428173.2 |
NRK
|
Nik related kinase |
| chr5_-_10249990 | 0.23 |
ENST00000511437.1
ENST00000280330.8 ENST00000510047.1 |
FAM173B
|
family with sequence similarity 173, member B |
| chr12_+_7053172 | 0.23 |
ENST00000229281.5
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr2_-_87248975 | 0.23 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr14_+_60558627 | 0.23 |
ENST00000317623.4
ENST00000391611.2 ENST00000406854.1 ENST00000406949.1 |
PCNXL4
|
pecanex-like 4 (Drosophila) |
| chr4_-_122791583 | 0.23 |
ENST00000506636.1
ENST00000264499.4 |
BBS7
|
Bardet-Biedl syndrome 7 |
| chr1_+_236686875 | 0.22 |
ENST00000366584.4
|
LGALS8
|
lectin, galactoside-binding, soluble, 8 |
| chr3_+_32737027 | 0.22 |
ENST00000454516.2
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr3_+_49977523 | 0.22 |
ENST00000422955.1
|
RBM6
|
RNA binding motif protein 6 |
| chr13_+_32889605 | 0.22 |
ENST00000380152.3
ENST00000544455.1 ENST00000530893.2 |
BRCA2
|
breast cancer 2, early onset |
| chr17_+_39394250 | 0.22 |
ENST00000254072.6
|
KRTAP9-8
|
keratin associated protein 9-8 |
| chr16_-_14724057 | 0.22 |
ENST00000539279.1
ENST00000420015.2 ENST00000437198.2 |
PARN
|
poly(A)-specific ribonuclease |
| chr2_-_178483694 | 0.22 |
ENST00000355689.5
|
TTC30A
|
tetratricopeptide repeat domain 30A |
| chr17_-_9479128 | 0.21 |
ENST00000574431.1
|
STX8
|
syntaxin 8 |
| chr6_-_116601044 | 0.21 |
ENST00000368608.3
|
TSPYL1
|
TSPY-like 1 |
| chr14_+_21387508 | 0.21 |
ENST00000555624.1
|
RP11-84C10.2
|
RP11-84C10.2 |
| chr3_+_49711777 | 0.21 |
ENST00000442186.1
ENST00000438011.1 ENST00000457042.1 |
APEH
|
acylaminoacyl-peptide hydrolase |
| chr19_-_36643329 | 0.21 |
ENST00000589154.1
|
COX7A1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr19_-_40023450 | 0.20 |
ENST00000326282.4
|
EID2B
|
EP300 interacting inhibitor of differentiation 2B |
| chr12_-_123187890 | 0.20 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr7_+_23221613 | 0.20 |
ENST00000410002.3
ENST00000413919.1 |
NUPL2
|
nucleoporin like 2 |
| chr9_+_71394945 | 0.20 |
ENST00000394264.3
|
FAM122A
|
family with sequence similarity 122A |
| chr12_+_51318513 | 0.20 |
ENST00000332160.4
|
METTL7A
|
methyltransferase like 7A |
| chr15_-_63449663 | 0.20 |
ENST00000439025.1
|
RPS27L
|
ribosomal protein S27-like |
| chr11_+_7110165 | 0.19 |
ENST00000306904.5
|
RBMXL2
|
RNA binding motif protein, X-linked-like 2 |
| chr2_+_113033164 | 0.19 |
ENST00000409871.1
ENST00000343936.4 |
ZC3H6
|
zinc finger CCCH-type containing 6 |
| chr12_+_69753448 | 0.19 |
ENST00000247843.2
ENST00000548020.1 ENST00000549685.1 ENST00000552955.1 |
YEATS4
|
YEATS domain containing 4 |
| chr17_-_7387524 | 0.19 |
ENST00000311403.4
|
ZBTB4
|
zinc finger and BTB domain containing 4 |
| chr9_+_131644388 | 0.19 |
ENST00000372600.4
|
LRRC8A
|
leucine rich repeat containing 8 family, member A |
| chr1_+_180601139 | 0.19 |
ENST00000367590.4
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr22_+_22712087 | 0.18 |
ENST00000390294.2
|
IGLV1-47
|
immunoglobulin lambda variable 1-47 |
| chr12_+_7052974 | 0.18 |
ENST00000544681.1
ENST00000537087.1 |
C12orf57
|
chromosome 12 open reading frame 57 |
| chr4_+_119200215 | 0.18 |
ENST00000602573.1
|
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr7_-_100034060 | 0.18 |
ENST00000292330.2
|
PPP1R35
|
protein phosphatase 1, regulatory subunit 35 |
| chr12_-_76478686 | 0.18 |
ENST00000261182.8
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chrX_-_153637612 | 0.18 |
ENST00000369807.1
ENST00000369808.3 |
DNASE1L1
|
deoxyribonuclease I-like 1 |
| chr3_-_122746628 | 0.18 |
ENST00000357599.3
|
SEMA5B
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5B |
| chr19_+_53970970 | 0.18 |
ENST00000468450.1
ENST00000396403.4 ENST00000490956.1 ENST00000396421.4 |
ZNF813
|
zinc finger protein 813 |
| chr21_-_40033618 | 0.18 |
ENST00000417133.2
ENST00000398910.1 ENST00000442448.1 |
ERG
|
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr9_+_131644398 | 0.18 |
ENST00000372599.3
|
LRRC8A
|
leucine rich repeat containing 8 family, member A |
| chr6_-_133119668 | 0.18 |
ENST00000275227.4
ENST00000538764.1 |
SLC18B1
|
solute carrier family 18, subfamily B, member 1 |
| chr2_-_191115229 | 0.18 |
ENST00000409820.2
ENST00000410045.1 |
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr6_-_29343068 | 0.18 |
ENST00000396806.3
|
OR12D3
|
olfactory receptor, family 12, subfamily D, member 3 |
| chr6_+_71377473 | 0.18 |
ENST00000370452.3
ENST00000316999.5 ENST00000370455.3 |
SMAP1
|
small ArfGAP 1 |
| chr19_-_12595586 | 0.18 |
ENST00000397732.3
|
ZNF709
|
zinc finger protein 709 |
| chr12_-_104359475 | 0.18 |
ENST00000553183.1
|
C12orf73
|
chromosome 12 open reading frame 73 |
| chr8_-_117768023 | 0.17 |
ENST00000518949.1
ENST00000522453.1 ENST00000521861.1 ENST00000518995.1 |
EIF3H
|
eukaryotic translation initiation factor 3, subunit H |
| chr1_+_233086326 | 0.17 |
ENST00000366628.5
ENST00000366627.4 |
NTPCR
|
nucleoside-triphosphatase, cancer-related |
| chr4_-_147443043 | 0.17 |
ENST00000394059.4
ENST00000502607.1 ENST00000335472.7 ENST00000432059.2 ENST00000394062.3 |
SLC10A7
|
solute carrier family 10, member 7 |
| chr12_+_133758115 | 0.17 |
ENST00000541009.2
ENST00000592241.1 |
ZNF268
|
zinc finger protein 268 |
| chr17_-_7166500 | 0.17 |
ENST00000575313.1
ENST00000397317.4 |
CLDN7
|
claudin 7 |
| chr19_+_57640011 | 0.17 |
ENST00000598197.1
|
USP29
|
ubiquitin specific peptidase 29 |
| chr1_-_19615744 | 0.17 |
ENST00000361640.4
|
AKR7A3
|
aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) |
| chr16_-_30569584 | 0.16 |
ENST00000252797.2
ENST00000568114.1 |
ZNF764
AC002310.13
|
zinc finger protein 764 Uncharacterized protein |
| chr19_-_9903666 | 0.16 |
ENST00000592587.1
|
ZNF846
|
zinc finger protein 846 |
| chr11_-_124981475 | 0.16 |
ENST00000532156.1
ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chr2_+_172864490 | 0.16 |
ENST00000315796.4
|
METAP1D
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr5_-_16179884 | 0.16 |
ENST00000332432.8
|
MARCH11
|
membrane-associated ring finger (C3HC4) 11 |
| chr8_+_96037255 | 0.16 |
ENST00000286687.4
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr14_-_58893832 | 0.16 |
ENST00000556007.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr2_+_133874577 | 0.16 |
ENST00000596384.1
|
AC011755.1
|
HCG2006742; Protein LOC100996685 |
| chr19_+_40502938 | 0.16 |
ENST00000599504.1
ENST00000596894.1 ENST00000601138.1 ENST00000600094.1 ENST00000347077.4 |
ZNF546
|
zinc finger protein 546 |
| chr2_+_33661382 | 0.16 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr19_+_58694396 | 0.16 |
ENST00000326804.4
ENST00000345813.3 ENST00000424679.2 |
ZNF274
|
zinc finger protein 274 |
| chr7_+_39605966 | 0.16 |
ENST00000223273.2
ENST00000448268.1 ENST00000432096.2 |
YAE1D1
|
Yae1 domain containing 1 |
| chr14_-_58894223 | 0.16 |
ENST00000555593.1
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr14_-_50101931 | 0.16 |
ENST00000298292.8
ENST00000406043.3 |
DNAAF2
|
dynein, axonemal, assembly factor 2 |
| chr8_+_105235572 | 0.16 |
ENST00000523362.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr8_+_25316489 | 0.16 |
ENST00000330560.3
|
CDCA2
|
cell division cycle associated 2 |
| chr2_-_55647057 | 0.16 |
ENST00000436346.1
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr8_+_42396936 | 0.16 |
ENST00000416469.2
|
SMIM19
|
small integral membrane protein 19 |
| chr13_-_37633743 | 0.15 |
ENST00000497318.1
ENST00000475892.1 ENST00000356185.3 ENST00000350612.6 ENST00000542180.1 ENST00000360252.4 |
SUPT20H
|
suppressor of Ty 20 homolog (S. cerevisiae) |
| chr7_+_80275621 | 0.15 |
ENST00000426978.1
ENST00000432207.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr8_+_25316707 | 0.15 |
ENST00000380665.3
|
CDCA2
|
cell division cycle associated 2 |
| chr13_-_37633567 | 0.15 |
ENST00000464744.1
|
SUPT20H
|
suppressor of Ty 20 homolog (S. cerevisiae) |
| chr4_+_147096837 | 0.15 |
ENST00000296581.5
ENST00000502781.1 |
LSM6
|
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr2_+_9563697 | 0.15 |
ENST00000238112.3
|
CPSF3
|
cleavage and polyadenylation specific factor 3, 73kDa |
| chrY_+_15815447 | 0.15 |
ENST00000284856.3
|
TMSB4Y
|
thymosin beta 4, Y-linked |
| chr5_-_40835303 | 0.14 |
ENST00000509877.1
ENST00000508493.1 ENST00000274242.5 |
RPL37
|
ribosomal protein L37 |
| chr16_+_89696692 | 0.14 |
ENST00000261615.4
|
DPEP1
|
dipeptidase 1 (renal) |
| chr14_+_58894103 | 0.14 |
ENST00000354386.6
ENST00000556134.1 |
KIAA0586
|
KIAA0586 |
| chr7_-_148581360 | 0.14 |
ENST00000320356.2
ENST00000541220.1 ENST00000483967.1 ENST00000536783.1 |
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr14_-_75518129 | 0.14 |
ENST00000556257.1
ENST00000557648.1 ENST00000553263.1 ENST00000355774.2 ENST00000380968.2 ENST00000238662.7 |
MLH3
|
mutL homolog 3 |
| chr19_+_24269981 | 0.14 |
ENST00000339642.6
ENST00000357002.4 |
ZNF254
|
zinc finger protein 254 |
| chr18_-_70211691 | 0.14 |
ENST00000269503.4
|
CBLN2
|
cerebellin 2 precursor |
| chr6_-_85473156 | 0.14 |
ENST00000606784.1
ENST00000606325.1 |
TBX18
|
T-box 18 |
| chr2_-_70475586 | 0.14 |
ENST00000416149.2
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr7_-_38948774 | 0.13 |
ENST00000395969.2
ENST00000414632.1 ENST00000310301.4 |
VPS41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr2_-_55646957 | 0.13 |
ENST00000263630.8
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr14_-_31889782 | 0.13 |
ENST00000543095.2
|
HEATR5A
|
HEAT repeat containing 5A |
| chrX_+_49969405 | 0.13 |
ENST00000376042.1
|
CCNB3
|
cyclin B3 |
| chr11_+_122753391 | 0.13 |
ENST00000307257.6
ENST00000227349.2 |
C11orf63
|
chromosome 11 open reading frame 63 |
| chr14_+_57857262 | 0.13 |
ENST00000555166.1
ENST00000556492.1 ENST00000554703.1 |
NAA30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chr12_-_62997214 | 0.13 |
ENST00000408887.2
|
C12orf61
|
chromosome 12 open reading frame 61 |
| chr3_+_134205000 | 0.13 |
ENST00000512894.1
ENST00000513612.2 ENST00000606977.1 |
CEP63
|
centrosomal protein 63kDa |
| chr6_+_43395272 | 0.13 |
ENST00000372530.4
|
ABCC10
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 10 |
| chr7_-_124569991 | 0.13 |
ENST00000446993.1
ENST00000357628.3 ENST00000393329.1 |
POT1
|
protection of telomeres 1 |
| chr9_+_4679555 | 0.12 |
ENST00000381858.1
ENST00000381854.3 |
CDC37L1
|
cell division cycle 37-like 1 |
| chr7_-_112758589 | 0.12 |
ENST00000413744.1
ENST00000439551.1 ENST00000441359.1 |
LINC00998
|
long intergenic non-protein coding RNA 998 |
| chr12_-_25055177 | 0.12 |
ENST00000538118.1
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr1_-_211848899 | 0.12 |
ENST00000366998.3
ENST00000540251.1 ENST00000366999.4 |
NEK2
|
NIMA-related kinase 2 |
| chr3_+_160117418 | 0.12 |
ENST00000465903.1
ENST00000485645.1 ENST00000360111.2 ENST00000472991.1 ENST00000467468.1 ENST00000469762.1 ENST00000489573.1 ENST00000462787.1 ENST00000490207.1 ENST00000485867.1 |
SMC4
|
structural maintenance of chromosomes 4 |
| chr16_-_30569801 | 0.12 |
ENST00000395091.2
|
ZNF764
|
zinc finger protein 764 |
| chr3_+_52489606 | 0.12 |
ENST00000488380.1
ENST00000420808.2 |
NISCH
|
nischarin |
| chr11_-_78285804 | 0.12 |
ENST00000281038.5
ENST00000529571.1 |
NARS2
|
asparaginyl-tRNA synthetase 2, mitochondrial (putative) |
| chrX_-_135962876 | 0.12 |
ENST00000431446.3
ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX
|
RNA binding motif protein, X-linked |
| chr14_+_21387491 | 0.12 |
ENST00000258817.2
|
RP11-84C10.2
|
RP11-84C10.2 |
| chr4_+_17812525 | 0.12 |
ENST00000251496.2
|
NCAPG
|
non-SMC condensin I complex, subunit G |
| chr5_-_147211190 | 0.12 |
ENST00000510027.2
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1 |
| chr1_+_53662101 | 0.12 |
ENST00000371486.3
|
CPT2
|
carnitine palmitoyltransferase 2 |
| chr10_-_14590644 | 0.12 |
ENST00000378470.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr11_+_118938485 | 0.12 |
ENST00000300793.6
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae) |
| chr4_+_79472673 | 0.12 |
ENST00000264908.6
|
ANXA3
|
annexin A3 |
| chr19_-_58485895 | 0.12 |
ENST00000314391.3
|
C19orf18
|
chromosome 19 open reading frame 18 |
| chr5_-_154317740 | 0.11 |
ENST00000285873.7
|
GEMIN5
|
gem (nuclear organelle) associated protein 5 |
| chr11_+_57365150 | 0.11 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr1_+_40840320 | 0.11 |
ENST00000372708.1
|
SMAP2
|
small ArfGAP2 |
| chr3_+_134204881 | 0.11 |
ENST00000511574.1
ENST00000337090.3 ENST00000383229.3 |
CEP63
|
centrosomal protein 63kDa |
| chr12_+_11802753 | 0.11 |
ENST00000396373.4
|
ETV6
|
ets variant 6 |
| chr2_+_190541153 | 0.11 |
ENST00000313581.4
ENST00000438402.2 ENST00000431575.2 ENST00000281412.6 |
ANKAR
|
ankyrin and armadillo repeat containing |
Network of associatons between targets according to the STRING database.
First level regulatory network of MYBL1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.2 | 0.5 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.2 | 0.5 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.3 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.1 | 0.7 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.1 | 0.7 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.1 | 0.3 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.1 | 0.2 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.1 | 0.5 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.1 | 0.5 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.3 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.1 | 0.3 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.4 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 | 0.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.4 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.1 | 0.2 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.6 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 0.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.2 | GO:1901535 | regulation of DNA demethylation(GO:1901535) |
| 0.0 | 0.3 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.1 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.0 | 0.1 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.0 | 0.3 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) |
| 0.0 | 0.2 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.0 | 0.2 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.2 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.4 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.3 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.4 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.3 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:0046080 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.0 | 0.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.2 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.3 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.3 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.1 | GO:0051311 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.3 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.3 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.2 | GO:0070162 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.0 | 0.3 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.1 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.1 | GO:0018282 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.0 | 0.1 | GO:0051039 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 0.6 | GO:1905145 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 0.2 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.2 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.2 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.4 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.1 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.4 | GO:0006525 | arginine metabolic process(GO:0006525) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.0 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.0 | 0.0 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.0 | 1.2 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0051096 | telomere assembly(GO:0032202) positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.0 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.0 | GO:0071048 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.0 | 0.1 | GO:0021649 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.3 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 0.5 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.2 | GO:0005712 | chiasma(GO:0005712) |
| 0.1 | 0.4 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.1 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.0 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.0 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.7 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 0.5 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.3 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.1 | 0.7 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.5 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.6 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 0.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.3 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.2 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.0 | 0.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.3 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0031071 | cysteine desulfurase activity(GO:0031071) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:0043337 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.5 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.0 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) centromeric DNA binding(GO:0019237) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.0 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.0 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.2 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.9 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.9 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |