Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
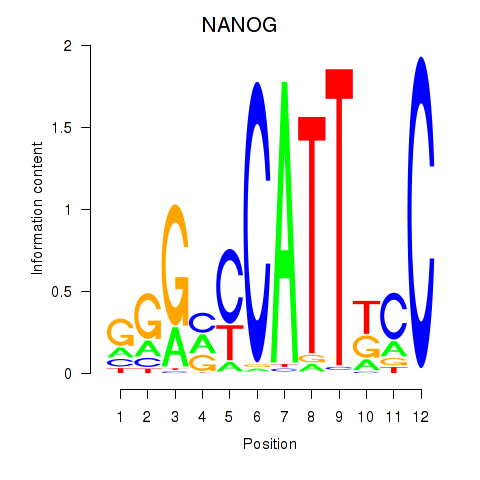
Results for NANOG
Z-value: 0.46
Transcription factors associated with NANOG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NANOG
|
ENSG00000111704.6 | Nanog homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NANOG | hg19_v2_chr12_+_7941989_7942014 | -0.63 | 8.3e-04 | Click! |
Activity profile of NANOG motif
Sorted Z-values of NANOG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_16900184 | 1.82 |
ENST00000515064.1
|
LDB2
|
LIM domain binding 2 |
| chr4_-_16900217 | 1.80 |
ENST00000441778.2
|
LDB2
|
LIM domain binding 2 |
| chr4_-_16900242 | 1.72 |
ENST00000502640.1
ENST00000506732.1 |
LDB2
|
LIM domain binding 2 |
| chr4_-_16900410 | 1.48 |
ENST00000304523.5
|
LDB2
|
LIM domain binding 2 |
| chr12_-_123201337 | 1.16 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr3_-_93692781 | 0.95 |
ENST00000394236.3
|
PROS1
|
protein S (alpha) |
| chr3_+_32147997 | 0.88 |
ENST00000282541.5
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr15_-_78526855 | 0.83 |
ENST00000541759.1
ENST00000558130.1 |
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr12_-_123187890 | 0.75 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr1_+_210406121 | 0.71 |
ENST00000367012.3
|
SERTAD4
|
SERTA domain containing 4 |
| chr6_-_119031228 | 0.61 |
ENST00000392500.3
ENST00000368488.5 ENST00000434604.1 |
CEP85L
|
centrosomal protein 85kDa-like |
| chr12_-_53901266 | 0.52 |
ENST00000609999.1
ENST00000267017.3 |
NPFF
|
neuropeptide FF-amide peptide precursor |
| chr2_+_45878790 | 0.48 |
ENST00000306156.3
|
PRKCE
|
protein kinase C, epsilon |
| chr7_+_150382781 | 0.47 |
ENST00000223293.5
ENST00000474605.1 |
GIMAP2
|
GTPase, IMAP family member 2 |
| chr6_+_155537771 | 0.46 |
ENST00000275246.7
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr19_-_13213662 | 0.44 |
ENST00000264824.4
|
LYL1
|
lymphoblastic leukemia derived sequence 1 |
| chr10_+_129845823 | 0.44 |
ENST00000306042.5
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E |
| chr2_+_87769459 | 0.43 |
ENST00000414030.1
ENST00000437561.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr6_+_148663729 | 0.42 |
ENST00000367467.3
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr2_-_240322643 | 0.41 |
ENST00000345617.3
|
HDAC4
|
histone deacetylase 4 |
| chr19_-_42806919 | 0.41 |
ENST00000595530.1
ENST00000538771.1 ENST00000601865.1 |
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr12_-_25055177 | 0.41 |
ENST00000538118.1
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr14_+_21156915 | 0.39 |
ENST00000397990.4
ENST00000555597.1 |
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5 ribonuclease, RNase A family, 4 |
| chr2_-_112237835 | 0.38 |
ENST00000442293.1
ENST00000439494.1 |
MIR4435-1HG
|
MIR4435-1 host gene (non-protein coding) |
| chr13_+_108922228 | 0.38 |
ENST00000542136.1
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr8_-_27469196 | 0.37 |
ENST00000546343.1
ENST00000560566.1 |
CLU
|
clusterin |
| chr19_-_42806723 | 0.36 |
ENST00000262890.3
|
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr3_-_49851313 | 0.35 |
ENST00000333486.3
|
UBA7
|
ubiquitin-like modifier activating enzyme 7 |
| chr8_-_27468842 | 0.35 |
ENST00000523500.1
|
CLU
|
clusterin |
| chr19_-_42806444 | 0.35 |
ENST00000594989.1
|
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr8_-_27468945 | 0.34 |
ENST00000405140.3
|
CLU
|
clusterin |
| chr1_-_147245484 | 0.33 |
ENST00000271348.2
|
GJA5
|
gap junction protein, alpha 5, 40kDa |
| chr14_-_98444438 | 0.33 |
ENST00000512901.2
|
C14orf64
|
chromosome 14 open reading frame 64 |
| chr6_+_26087509 | 0.33 |
ENST00000397022.3
ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE
|
hemochromatosis |
| chr17_+_7387677 | 0.31 |
ENST00000322644.6
|
POLR2A
|
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa |
| chr3_-_149093499 | 0.30 |
ENST00000472441.1
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr16_+_31494323 | 0.29 |
ENST00000569576.1
ENST00000330498.3 |
SLC5A2
|
solute carrier family 5 (sodium/glucose cotransporter), member 2 |
| chr4_-_16077741 | 0.28 |
ENST00000447510.2
ENST00000540805.1 ENST00000539194.1 |
PROM1
|
prominin 1 |
| chr17_-_34417479 | 0.27 |
ENST00000225245.5
|
CCL3
|
chemokine (C-C motif) ligand 3 |
| chr19_-_42806842 | 0.26 |
ENST00000596265.1
|
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr10_+_69644404 | 0.26 |
ENST00000212015.6
|
SIRT1
|
sirtuin 1 |
| chr17_-_7387524 | 0.24 |
ENST00000311403.4
|
ZBTB4
|
zinc finger and BTB domain containing 4 |
| chr1_-_205649580 | 0.24 |
ENST00000367145.3
|
SLC45A3
|
solute carrier family 45, member 3 |
| chr3_+_132379154 | 0.24 |
ENST00000468022.1
ENST00000473651.1 ENST00000494238.2 |
UBA5
|
ubiquitin-like modifier activating enzyme 5 |
| chr16_+_68573640 | 0.24 |
ENST00000398253.2
ENST00000573161.1 |
ZFP90
|
ZFP90 zinc finger protein |
| chr12_-_31743901 | 0.24 |
ENST00000354285.4
|
DENND5B
|
DENN/MADD domain containing 5B |
| chr3_+_107244229 | 0.23 |
ENST00000456419.1
ENST00000402163.2 |
BBX
|
bobby sox homolog (Drosophila) |
| chr1_-_47655686 | 0.23 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr18_+_13218769 | 0.23 |
ENST00000399848.3
ENST00000361205.4 |
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr16_+_640055 | 0.23 |
ENST00000568586.1
ENST00000538492.1 ENST00000248139.3 |
RAB40C
|
RAB40C, member RAS oncogene family |
| chrX_-_107019181 | 0.22 |
ENST00000315660.4
ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr19_+_42381173 | 0.22 |
ENST00000221972.3
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr20_+_37554955 | 0.22 |
ENST00000217429.4
|
FAM83D
|
family with sequence similarity 83, member D |
| chr2_-_97405775 | 0.22 |
ENST00000264963.4
ENST00000537039.1 ENST00000377079.4 ENST00000426463.2 ENST00000534882.1 |
LMAN2L
|
lectin, mannose-binding 2-like |
| chrX_-_118827333 | 0.21 |
ENST00000360156.7
ENST00000354228.4 ENST00000489216.1 ENST00000354416.3 ENST00000394610.1 ENST00000343984.5 |
SEPT6
|
septin 6 |
| chrX_-_133931164 | 0.21 |
ENST00000370790.1
ENST00000298090.6 |
FAM122B
|
family with sequence similarity 122B |
| chr4_+_166248775 | 0.21 |
ENST00000261507.6
ENST00000507013.1 ENST00000393766.2 ENST00000504317.1 |
MSMO1
|
methylsterol monooxygenase 1 |
| chr2_+_170590321 | 0.20 |
ENST00000392647.2
|
KLHL23
|
kelch-like family member 23 |
| chr17_-_72542278 | 0.20 |
ENST00000330793.1
|
CD300C
|
CD300c molecule |
| chr1_-_202129105 | 0.20 |
ENST00000367279.4
|
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr14_+_67999999 | 0.20 |
ENST00000329153.5
|
PLEKHH1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
| chr6_+_90272027 | 0.19 |
ENST00000522441.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr2_-_69664586 | 0.19 |
ENST00000303698.3
ENST00000394305.1 ENST00000410022.2 |
NFU1
|
NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) |
| chr19_-_54327542 | 0.19 |
ENST00000391775.3
ENST00000324134.6 ENST00000535162.1 ENST00000351894.4 ENST00000354278.3 ENST00000391773.1 ENST00000345770.5 ENST00000391772.1 |
NLRP12
|
NLR family, pyrin domain containing 12 |
| chr18_+_3262954 | 0.19 |
ENST00000584539.1
|
MYL12B
|
myosin, light chain 12B, regulatory |
| chr1_-_202129704 | 0.18 |
ENST00000476061.1
ENST00000544762.1 ENST00000467283.1 ENST00000464870.1 ENST00000435759.2 ENST00000486116.1 ENST00000543735.1 ENST00000308986.5 ENST00000477625.1 |
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr7_+_102715315 | 0.18 |
ENST00000428183.2
ENST00000323716.3 ENST00000441711.2 ENST00000454559.1 ENST00000425331.1 ENST00000541300.1 |
ARMC10
|
armadillo repeat containing 10 |
| chr6_-_20212630 | 0.18 |
ENST00000324607.7
ENST00000541730.1 ENST00000536798.1 |
MBOAT1
|
membrane bound O-acyltransferase domain containing 1 |
| chr16_+_68573116 | 0.18 |
ENST00000570495.1
ENST00000563169.2 ENST00000564323.1 ENST00000562156.1 ENST00000573685.1 |
ZFP90
|
ZFP90 zinc finger protein |
| chr17_+_43224684 | 0.18 |
ENST00000332499.2
|
HEXIM1
|
hexamethylene bis-acetamide inducible 1 |
| chr6_-_107077347 | 0.17 |
ENST00000369063.3
ENST00000539449.1 |
RTN4IP1
|
reticulon 4 interacting protein 1 |
| chr3_-_134369853 | 0.17 |
ENST00000508956.1
ENST00000503669.1 ENST00000423778.2 |
KY
|
kyphoscoliosis peptidase |
| chr17_+_75372165 | 0.17 |
ENST00000427674.2
|
SEPT9
|
septin 9 |
| chrX_+_103173457 | 0.17 |
ENST00000419165.1
|
TMSB15B
|
thymosin beta 15B |
| chr6_-_33714667 | 0.17 |
ENST00000293756.4
|
IP6K3
|
inositol hexakisphosphate kinase 3 |
| chr6_+_292051 | 0.17 |
ENST00000344450.5
|
DUSP22
|
dual specificity phosphatase 22 |
| chr1_+_159272111 | 0.16 |
ENST00000368114.1
|
FCER1A
|
Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide |
| chr1_+_27719148 | 0.16 |
ENST00000374024.3
|
GPR3
|
G protein-coupled receptor 3 |
| chr16_-_70719925 | 0.16 |
ENST00000338779.6
|
MTSS1L
|
metastasis suppressor 1-like |
| chr1_+_47264711 | 0.15 |
ENST00000371923.4
ENST00000271153.4 ENST00000371919.4 |
CYP4B1
|
cytochrome P450, family 4, subfamily B, polypeptide 1 |
| chr9_-_99381660 | 0.15 |
ENST00000375240.3
ENST00000463569.1 |
CDC14B
|
cell division cycle 14B |
| chr1_+_161551101 | 0.15 |
ENST00000367962.4
ENST00000367960.5 ENST00000403078.3 ENST00000428605.2 |
FCGR2B
|
Fc fragment of IgG, low affinity IIb, receptor (CD32) |
| chr6_-_33714752 | 0.15 |
ENST00000451316.1
|
IP6K3
|
inositol hexakisphosphate kinase 3 |
| chr7_+_131012605 | 0.14 |
ENST00000446815.1
ENST00000352689.6 |
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr6_+_151561085 | 0.14 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr11_-_67205538 | 0.14 |
ENST00000326294.3
|
PTPRCAP
|
protein tyrosine phosphatase, receptor type, C-associated protein |
| chr20_-_3762087 | 0.14 |
ENST00000379756.3
|
SPEF1
|
sperm flagellar 1 |
| chr22_+_31090793 | 0.13 |
ENST00000332585.6
ENST00000382310.3 ENST00000446658.2 |
OSBP2
|
oxysterol binding protein 2 |
| chr22_+_41487711 | 0.13 |
ENST00000263253.7
|
EP300
|
E1A binding protein p300 |
| chr1_+_173837214 | 0.13 |
ENST00000367704.1
|
ZBTB37
|
zinc finger and BTB domain containing 37 |
| chr17_+_7905912 | 0.13 |
ENST00000254854.4
|
GUCY2D
|
guanylate cyclase 2D, membrane (retina-specific) |
| chr14_+_23842018 | 0.13 |
ENST00000397242.2
ENST00000329715.2 |
IL25
|
interleukin 25 |
| chr15_+_44580955 | 0.12 |
ENST00000345795.2
ENST00000360824.3 |
CASC4
|
cancer susceptibility candidate 4 |
| chr19_+_51273721 | 0.11 |
ENST00000270590.4
|
GPR32
|
G protein-coupled receptor 32 |
| chr4_+_154073469 | 0.11 |
ENST00000441616.1
|
TRIM2
|
tripartite motif containing 2 |
| chr12_+_69080734 | 0.11 |
ENST00000378905.2
|
NUP107
|
nucleoporin 107kDa |
| chr6_+_26087646 | 0.11 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chr22_-_42084863 | 0.10 |
ENST00000401959.1
ENST00000355257.3 |
NHP2L1
|
NHP2 non-histone chromosome protein 2-like 1 (S. cerevisiae) |
| chr15_+_44580899 | 0.10 |
ENST00000559222.1
ENST00000299957.6 |
CASC4
|
cancer susceptibility candidate 4 |
| chr6_-_43021437 | 0.10 |
ENST00000265348.3
|
CUL7
|
cullin 7 |
| chr1_+_161632937 | 0.10 |
ENST00000236937.9
ENST00000367961.4 ENST00000358671.5 |
FCGR2B
|
Fc fragment of IgG, low affinity IIb, receptor (CD32) |
| chr15_+_78832747 | 0.10 |
ENST00000560217.1
ENST00000044462.7 ENST00000559082.1 ENST00000559948.1 ENST00000413382.2 ENST00000559146.1 ENST00000558281.1 |
PSMA4
|
proteasome (prosome, macropain) subunit, alpha type, 4 |
| chr1_-_145589424 | 0.10 |
ENST00000334513.5
|
NUDT17
|
nudix (nucleoside diphosphate linked moiety X)-type motif 17 |
| chrX_+_44703249 | 0.10 |
ENST00000339042.4
|
DUSP21
|
dual specificity phosphatase 21 |
| chrX_+_47092314 | 0.09 |
ENST00000218348.3
|
USP11
|
ubiquitin specific peptidase 11 |
| chr19_+_50084561 | 0.09 |
ENST00000246794.5
|
PRRG2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr8_+_9413410 | 0.09 |
ENST00000520408.1
ENST00000310430.6 ENST00000522110.1 |
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr22_-_30662828 | 0.09 |
ENST00000403463.1
ENST00000215781.2 |
OSM
|
oncostatin M |
| chr19_+_6372444 | 0.09 |
ENST00000245812.3
|
ALKBH7
|
alkB, alkylation repair homolog 7 (E. coli) |
| chr3_-_98241358 | 0.09 |
ENST00000503004.1
ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1
|
claudin domain containing 1 |
| chr12_-_57410304 | 0.09 |
ENST00000441881.1
ENST00000458521.2 |
TAC3
|
tachykinin 3 |
| chr6_+_52226897 | 0.09 |
ENST00000442253.2
|
PAQR8
|
progestin and adipoQ receptor family member VIII |
| chr16_-_75301886 | 0.09 |
ENST00000393422.2
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr19_+_15197791 | 0.08 |
ENST00000209540.2
|
OR1I1
|
olfactory receptor, family 1, subfamily I, member 1 |
| chr8_-_91095099 | 0.08 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chrX_+_134124968 | 0.08 |
ENST00000330288.4
|
SMIM10
|
small integral membrane protein 10 |
| chr5_+_139493665 | 0.08 |
ENST00000331327.3
|
PURA
|
purine-rich element binding protein A |
| chr16_+_30759700 | 0.08 |
ENST00000328273.7
|
PHKG2
|
phosphorylase kinase, gamma 2 (testis) |
| chr10_-_70092671 | 0.08 |
ENST00000358769.2
ENST00000432941.1 ENST00000495025.2 |
PBLD
|
phenazine biosynthesis-like protein domain containing |
| chr6_-_43021612 | 0.07 |
ENST00000535468.1
|
CUL7
|
cullin 7 |
| chr2_-_198650037 | 0.07 |
ENST00000392296.4
|
BOLL
|
boule-like RNA-binding protein |
| chr3_-_52567792 | 0.07 |
ENST00000307092.4
ENST00000422318.2 ENST00000459839.1 |
NT5DC2
|
5'-nucleotidase domain containing 2 |
| chr10_+_5566916 | 0.06 |
ENST00000315238.1
|
CALML3
|
calmodulin-like 3 |
| chr20_+_36974759 | 0.06 |
ENST00000217407.2
|
LBP
|
lipopolysaccharide binding protein |
| chr5_-_55290773 | 0.06 |
ENST00000502326.3
ENST00000381298.2 |
IL6ST
|
interleukin 6 signal transducer (gp130, oncostatin M receptor) |
| chr16_+_640201 | 0.06 |
ENST00000563109.1
|
RAB40C
|
RAB40C, member RAS oncogene family |
| chr19_-_39926555 | 0.06 |
ENST00000599539.1
ENST00000339471.4 ENST00000601655.1 ENST00000251453.3 |
RPS16
|
ribosomal protein S16 |
| chr10_+_102891048 | 0.06 |
ENST00000467928.2
|
TLX1
|
T-cell leukemia homeobox 1 |
| chr18_+_3262415 | 0.06 |
ENST00000581193.1
ENST00000400175.5 |
MYL12B
|
myosin, light chain 12B, regulatory |
| chr8_-_42751820 | 0.06 |
ENST00000526349.1
ENST00000527424.1 ENST00000534961.1 ENST00000319073.4 |
RNF170
|
ring finger protein 170 |
| chrX_+_103357202 | 0.06 |
ENST00000537356.3
|
ZCCHC18
|
zinc finger, CCHC domain containing 18 |
| chr20_+_43343517 | 0.06 |
ENST00000372865.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr19_+_37825614 | 0.05 |
ENST00000591259.1
ENST00000590582.1 |
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr19_-_36870087 | 0.05 |
ENST00000270001.7
|
ZFP14
|
ZFP14 zinc finger protein |
| chr4_+_154074217 | 0.05 |
ENST00000437508.2
|
TRIM2
|
tripartite motif containing 2 |
| chr1_+_173837488 | 0.05 |
ENST00000427304.1
ENST00000432989.1 ENST00000367702.1 |
ZBTB37
|
zinc finger and BTB domain containing 37 |
| chr12_-_52946923 | 0.05 |
ENST00000267119.5
|
KRT71
|
keratin 71 |
| chr10_+_64133934 | 0.05 |
ENST00000395254.3
ENST00000395255.3 ENST00000410046.3 |
ZNF365
|
zinc finger protein 365 |
| chr20_+_43343476 | 0.04 |
ENST00000372868.2
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr10_-_1034237 | 0.04 |
ENST00000381466.1
|
AL359878.1
|
Uncharacterized protein |
| chr1_-_155959853 | 0.04 |
ENST00000462460.2
ENST00000368316.1 |
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr19_+_8455200 | 0.04 |
ENST00000601897.1
ENST00000594216.1 |
RAB11B
|
RAB11B, member RAS oncogene family |
| chr17_-_56606664 | 0.04 |
ENST00000580844.1
|
SEPT4
|
septin 4 |
| chr6_+_88032299 | 0.04 |
ENST00000608353.1
ENST00000392863.1 ENST00000229570.5 ENST00000608525.1 ENST00000608868.1 |
SMIM8
|
small integral membrane protein 8 |
| chr16_+_6069072 | 0.04 |
ENST00000547605.1
ENST00000550418.1 ENST00000553186.1 |
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr3_+_23847394 | 0.03 |
ENST00000306627.3
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chrX_+_85403445 | 0.03 |
ENST00000373131.1
|
DACH2
|
dachshund homolog 2 (Drosophila) |
| chr2_+_64681641 | 0.03 |
ENST00000409537.2
|
LGALSL
|
lectin, galactoside-binding-like |
| chr12_-_69080590 | 0.03 |
ENST00000433116.2
ENST00000500695.2 |
RP11-637A17.2
|
RP11-637A17.2 |
| chr6_-_136610911 | 0.03 |
ENST00000530767.1
ENST00000527759.1 ENST00000527536.1 ENST00000529826.1 ENST00000531224.1 ENST00000353331.4 |
BCLAF1
|
BCL2-associated transcription factor 1 |
| chr1_+_36348790 | 0.03 |
ENST00000373204.4
|
AGO1
|
argonaute RISC catalytic component 1 |
| chrX_-_120066151 | 0.03 |
ENST00000419982.2
ENST00000416816.1 |
CT47A12
|
cancer/testis antigen family 47, member A12 |
| chr6_-_131277510 | 0.03 |
ENST00000525193.1
ENST00000527659.1 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr15_+_73976715 | 0.03 |
ENST00000558689.1
ENST00000560786.2 ENST00000561213.1 ENST00000563584.1 ENST00000561416.1 |
CD276
|
CD276 molecule |
| chr19_-_39926268 | 0.02 |
ENST00000599705.1
|
RPS16
|
ribosomal protein S16 |
| chr20_+_43343886 | 0.02 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr22_+_26565440 | 0.02 |
ENST00000404234.3
ENST00000529632.2 ENST00000360929.3 ENST00000248933.6 ENST00000343706.4 |
SEZ6L
|
seizure related 6 homolog (mouse)-like |
| chr19_+_41816053 | 0.02 |
ENST00000269967.3
|
CCDC97
|
coiled-coil domain containing 97 |
| chr21_-_34960930 | 0.01 |
ENST00000437395.1
|
DONSON
|
downstream neighbor of SON |
| chr1_+_203097407 | 0.01 |
ENST00000367235.1
|
ADORA1
|
adenosine A1 receptor |
| chr4_+_123300121 | 0.01 |
ENST00000446706.1
ENST00000296513.2 |
ADAD1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chrX_-_120119638 | 0.01 |
ENST00000434414.1
ENST00000433030.1 ENST00000371280.3 |
CT47A1
|
cancer/testis antigen family 47, member A1 |
| chrX_+_85403487 | 0.01 |
ENST00000373125.4
|
DACH2
|
dachshund homolog 2 (Drosophila) |
| chr13_+_108921977 | 0.01 |
ENST00000430559.1
ENST00000375887.4 |
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr2_+_171673072 | 0.00 |
ENST00000358196.3
ENST00000375272.1 |
GAD1
|
glutamate decarboxylase 1 (brain, 67kDa) |
| chr12_-_123215306 | 0.00 |
ENST00000356987.2
ENST00000436083.2 |
HCAR1
|
hydroxycarboxylic acid receptor 1 |
| chr5_+_138089100 | 0.00 |
ENST00000520339.1
ENST00000355078.5 ENST00000302763.7 ENST00000518910.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr8_-_102181718 | 0.00 |
ENST00000565617.1
|
KB-1460A1.5
|
KB-1460A1.5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NANOG
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 | 6.8 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.2 | 1.1 | GO:0061518 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.2 | 0.5 | GO:0035669 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.1 | 0.4 | GO:1904437 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.1 | 0.5 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.1 | 0.8 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.3 | GO:0098904 | pulmonary valve formation(GO:0003193) regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) foramen ovale closure(GO:0035922) regulation of AV node cell action potential(GO:0098904) regulation of bundle of His cell action potential(GO:0098905) |
| 0.1 | 0.3 | GO:0001172 | transcription, RNA-templated(GO:0001172) |
| 0.1 | 0.3 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.1 | 0.4 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.2 | GO:0043318 | negative regulation of dendritic cell antigen processing and presentation(GO:0002605) regulation of cytotoxic T cell degranulation(GO:0043317) negative regulation of cytotoxic T cell degranulation(GO:0043318) |
| 0.1 | 0.2 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 0.4 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.1 | 0.3 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.1 | 0.4 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 | 0.4 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.9 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 0.2 | GO:0001812 | regulation of type I hypersensitivity(GO:0001810) positive regulation of type I hypersensitivity(GO:0001812) type I hypersensitivity(GO:0016068) |
| 0.0 | 0.2 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.4 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.3 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.4 | GO:0034983 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.2 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.2 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.7 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.2 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.1 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0009624 | response to nematode(GO:0009624) eosinophil differentiation(GO:0030222) |
| 0.0 | 0.1 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 1.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.1 | 0.4 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 0.2 | GO:0005940 | septin ring(GO:0005940) septin collar(GO:0032173) |
| 0.1 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.2 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 6.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 6.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.3 | 1.4 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.1 | 0.3 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.1 | 0.5 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.3 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.1 | 0.4 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.1 | 0.2 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.2 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.1 | 0.3 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.1 | 0.4 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.3 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.1 | 0.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.4 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.4 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 0.3 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 1.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.4 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) leukemia inhibitory factor receptor activity(GO:0004923) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.4 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.2 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |