Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for NFATC1
Z-value: 0.43
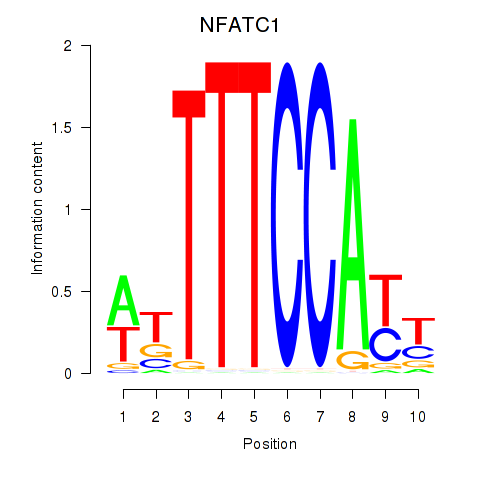
Transcription factors associated with NFATC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFATC1
|
ENSG00000131196.13 | nuclear factor of activated T cells 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFATC1 | hg19_v2_chr18_+_77155856_77155939 | 0.39 | 5.1e-02 | Click! |
Activity profile of NFATC1 motif
Sorted Z-values of NFATC1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_92413353 | 1.16 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr4_+_41614909 | 0.98 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr10_+_94590910 | 0.94 |
ENST00000371547.4
|
EXOC6
|
exocyst complex component 6 |
| chr21_-_35899113 | 0.75 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr1_+_61548225 | 0.58 |
ENST00000371187.3
|
NFIA
|
nuclear factor I/A |
| chr6_+_132891461 | 0.55 |
ENST00000275198.1
|
TAAR6
|
trace amine associated receptor 6 |
| chr13_+_32838801 | 0.53 |
ENST00000542859.1
|
FRY
|
furry homolog (Drosophila) |
| chr18_+_77155856 | 0.50 |
ENST00000253506.5
ENST00000591814.1 |
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr4_-_186733363 | 0.50 |
ENST00000393523.2
ENST00000393528.3 ENST00000449407.2 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_+_61547405 | 0.50 |
ENST00000371189.4
|
NFIA
|
nuclear factor I/A |
| chr14_-_92413727 | 0.45 |
ENST00000267620.10
|
FBLN5
|
fibulin 5 |
| chr9_-_14314518 | 0.45 |
ENST00000397581.2
|
NFIB
|
nuclear factor I/B |
| chr1_+_61548374 | 0.44 |
ENST00000485903.2
ENST00000371185.2 ENST00000371184.2 |
NFIA
|
nuclear factor I/A |
| chr9_-_14314566 | 0.43 |
ENST00000397579.2
|
NFIB
|
nuclear factor I/B |
| chr18_-_22932080 | 0.41 |
ENST00000584787.1
ENST00000361524.3 ENST00000538137.2 |
ZNF521
|
zinc finger protein 521 |
| chr21_-_27945562 | 0.39 |
ENST00000299340.4
ENST00000435845.2 |
CYYR1
|
cysteine/tyrosine-rich 1 |
| chr3_-_141747439 | 0.39 |
ENST00000467667.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr1_-_160924589 | 0.35 |
ENST00000368029.3
|
ITLN2
|
intelectin 2 |
| chr12_-_7848364 | 0.35 |
ENST00000329913.3
|
GDF3
|
growth differentiation factor 3 |
| chr14_+_63671577 | 0.32 |
ENST00000555125.1
|
RHOJ
|
ras homolog family member J |
| chr8_-_17555164 | 0.32 |
ENST00000297488.6
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr1_+_82266053 | 0.32 |
ENST00000370715.1
ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2
|
latrophilin 2 |
| chr18_+_77155942 | 0.32 |
ENST00000397790.2
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr9_-_16870704 | 0.31 |
ENST00000380672.4
ENST00000380667.2 ENST00000380666.2 ENST00000486514.1 |
BNC2
|
basonuclin 2 |
| chr3_-_141747459 | 0.28 |
ENST00000477292.1
ENST00000478006.1 ENST00000495310.1 ENST00000486111.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr12_+_93771659 | 0.28 |
ENST00000337179.5
ENST00000415493.2 |
NUDT4
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
| chr3_+_152017924 | 0.27 |
ENST00000465907.2
ENST00000492948.1 ENST00000485509.1 ENST00000464596.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chrX_+_41193407 | 0.26 |
ENST00000457138.2
ENST00000441189.2 |
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chrX_-_63005405 | 0.25 |
ENST00000374878.1
ENST00000437457.2 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr14_+_63671105 | 0.25 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr12_-_91573132 | 0.25 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr16_-_66952779 | 0.24 |
ENST00000570262.1
ENST00000394055.3 ENST00000299752.4 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr11_+_65190245 | 0.24 |
ENST00000499732.1
ENST00000501122.2 ENST00000601801.1 |
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
| chr5_+_135394840 | 0.24 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr1_+_66999799 | 0.23 |
ENST00000371035.3
ENST00000371036.3 ENST00000371037.4 |
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr3_+_25469724 | 0.23 |
ENST00000437042.2
|
RARB
|
retinoic acid receptor, beta |
| chr16_-_66952742 | 0.23 |
ENST00000565235.2
ENST00000568632.1 ENST00000565796.1 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr14_-_89883412 | 0.23 |
ENST00000557258.1
|
FOXN3
|
forkhead box N3 |
| chr12_-_50616122 | 0.23 |
ENST00000552823.1
ENST00000552909.1 |
LIMA1
|
LIM domain and actin binding 1 |
| chr5_-_111093759 | 0.22 |
ENST00000509979.1
ENST00000513100.1 ENST00000508161.1 ENST00000455559.2 |
NREP
|
neuronal regeneration related protein |
| chr5_-_88179302 | 0.22 |
ENST00000504921.2
|
MEF2C
|
myocyte enhancer factor 2C |
| chr12_-_50616382 | 0.21 |
ENST00000552783.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chrX_+_10031499 | 0.21 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chr4_-_159094194 | 0.21 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr10_-_72141330 | 0.21 |
ENST00000395011.1
ENST00000395010.1 |
LRRC20
|
leucine rich repeat containing 20 |
| chr19_+_41509851 | 0.20 |
ENST00000593831.1
ENST00000330446.5 |
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| chr12_+_49121213 | 0.20 |
ENST00000548380.1
|
LINC00935
|
long intergenic non-protein coding RNA 935 |
| chr11_+_73003824 | 0.20 |
ENST00000538328.1
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr12_+_96588279 | 0.20 |
ENST00000552142.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr3_-_185270342 | 0.19 |
ENST00000424591.2
|
LIPH
|
lipase, member H |
| chr12_+_27396901 | 0.19 |
ENST00000541191.1
ENST00000389032.3 |
STK38L
|
serine/threonine kinase 38 like |
| chr12_+_26348582 | 0.19 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chrX_+_152760397 | 0.19 |
ENST00000331595.4
ENST00000431891.1 |
BGN
|
biglycan |
| chr3_-_158390282 | 0.19 |
ENST00000264265.3
|
LXN
|
latexin |
| chr11_+_10477733 | 0.18 |
ENST00000528723.1
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr3_+_69134080 | 0.18 |
ENST00000273258.3
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr3_+_132316081 | 0.18 |
ENST00000249887.2
|
ACKR4
|
atypical chemokine receptor 4 |
| chr18_-_77276057 | 0.18 |
ENST00000597412.1
|
AC018445.1
|
Uncharacterized protein |
| chr12_-_92536433 | 0.18 |
ENST00000551563.2
ENST00000546975.1 ENST00000549802.1 |
C12orf79
|
chromosome 12 open reading frame 79 |
| chr15_-_28957469 | 0.17 |
ENST00000563027.1
ENST00000340249.3 |
GOLGA8M
|
golgin A8 family, member M |
| chr20_+_42574317 | 0.17 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2 |
| chr13_-_99667960 | 0.17 |
ENST00000448493.2
|
DOCK9
|
dedicator of cytokinesis 9 |
| chr15_+_100347228 | 0.17 |
ENST00000559714.1
ENST00000560059.1 |
CTD-2054N24.2
|
Uncharacterized protein |
| chr2_+_189839046 | 0.17 |
ENST00000304636.3
ENST00000317840.5 |
COL3A1
|
collagen, type III, alpha 1 |
| chr19_+_16059818 | 0.17 |
ENST00000322107.1
|
OR10H4
|
olfactory receptor, family 10, subfamily H, member 4 |
| chr4_-_104021009 | 0.16 |
ENST00000509245.1
ENST00000296424.4 |
BDH2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr18_+_3466248 | 0.16 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr8_+_86121448 | 0.16 |
ENST00000520225.1
|
E2F5
|
E2F transcription factor 5, p130-binding |
| chr8_+_81397846 | 0.16 |
ENST00000379091.4
|
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr15_-_43785274 | 0.16 |
ENST00000413546.1
|
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr15_-_74374891 | 0.16 |
ENST00000290438.3
|
GOLGA6A
|
golgin A6 family, member A |
| chr12_+_50366620 | 0.16 |
ENST00000315520.5
|
AQP6
|
aquaporin 6, kidney specific |
| chr18_-_53177984 | 0.15 |
ENST00000543082.1
|
TCF4
|
transcription factor 4 |
| chr2_+_11817713 | 0.15 |
ENST00000449576.2
|
LPIN1
|
lipin 1 |
| chr1_+_203734296 | 0.15 |
ENST00000442561.2
ENST00000367217.5 |
LAX1
|
lymphocyte transmembrane adaptor 1 |
| chr7_+_134551583 | 0.15 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr4_-_178363581 | 0.15 |
ENST00000264595.2
|
AGA
|
aspartylglucosaminidase |
| chr6_-_111136513 | 0.15 |
ENST00000368911.3
|
CDK19
|
cyclin-dependent kinase 19 |
| chr18_-_53257027 | 0.15 |
ENST00000568740.1
ENST00000564403.2 ENST00000537578.1 |
TCF4
|
transcription factor 4 |
| chr9_-_20622478 | 0.14 |
ENST00000355930.6
ENST00000380338.4 |
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr3_-_65583561 | 0.14 |
ENST00000460329.2
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr8_-_117043 | 0.14 |
ENST00000320901.3
|
OR4F21
|
olfactory receptor, family 4, subfamily F, member 21 |
| chr4_+_86748898 | 0.14 |
ENST00000509300.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr1_+_367640 | 0.14 |
ENST00000426406.1
|
OR4F29
|
olfactory receptor, family 4, subfamily F, member 29 |
| chr1_+_204042723 | 0.14 |
ENST00000367204.1
|
SOX13
|
SRY (sex determining region Y)-box 13 |
| chr18_-_52989217 | 0.14 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr6_+_15246501 | 0.14 |
ENST00000341776.2
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr9_+_22446814 | 0.14 |
ENST00000325870.2
|
DMRTA1
|
DMRT-like family A1 |
| chr8_-_81787006 | 0.14 |
ENST00000327835.3
|
ZNF704
|
zinc finger protein 704 |
| chr3_+_196366616 | 0.14 |
ENST00000426755.1
|
PIGX
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr3_-_149375783 | 0.14 |
ENST00000467467.1
ENST00000460517.1 ENST00000360632.3 |
WWTR1
|
WW domain containing transcription regulator 1 |
| chr19_+_15052301 | 0.14 |
ENST00000248072.3
|
OR7C2
|
olfactory receptor, family 7, subfamily C, member 2 |
| chr3_-_111852128 | 0.13 |
ENST00000308910.4
|
GCSAM
|
germinal center-associated, signaling and motility |
| chr11_+_11863500 | 0.13 |
ENST00000527733.1
ENST00000539466.1 |
USP47
|
ubiquitin specific peptidase 47 |
| chr5_+_140207536 | 0.13 |
ENST00000529310.1
ENST00000527624.1 |
PCDHA6
|
protocadherin alpha 6 |
| chr5_+_38445641 | 0.13 |
ENST00000397210.3
ENST00000506135.1 ENST00000508131.1 |
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains |
| chr4_+_154387480 | 0.13 |
ENST00000409663.3
ENST00000440693.1 ENST00000409959.3 |
KIAA0922
|
KIAA0922 |
| chr5_-_54468974 | 0.13 |
ENST00000381375.2
ENST00000296733.1 ENST00000322374.6 ENST00000334206.5 ENST00000331730.3 |
CDC20B
|
cell division cycle 20B |
| chr2_+_28618532 | 0.13 |
ENST00000545753.1
|
FOSL2
|
FOS-like antigen 2 |
| chr2_-_228244013 | 0.13 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr3_-_182817367 | 0.13 |
ENST00000265594.4
|
MCCC1
|
methylcrotonoyl-CoA carboxylase 1 (alpha) |
| chr9_-_4666337 | 0.13 |
ENST00000381890.5
|
SPATA6L
|
spermatogenesis associated 6-like |
| chr12_+_54393880 | 0.13 |
ENST00000303450.4
|
HOXC9
|
homeobox C9 |
| chr18_-_53089723 | 0.13 |
ENST00000561992.1
ENST00000562512.2 |
TCF4
|
transcription factor 4 |
| chr14_+_75761099 | 0.12 |
ENST00000561000.1
ENST00000558575.1 |
RP11-293M10.5
|
RP11-293M10.5 |
| chr11_-_68780824 | 0.12 |
ENST00000441623.1
ENST00000309099.6 |
MRGPRF
|
MAS-related GPR, member F |
| chr11_+_130542851 | 0.12 |
ENST00000317019.2
|
C11orf44
|
chromosome 11 open reading frame 44 |
| chr15_-_32695396 | 0.12 |
ENST00000512626.2
ENST00000435655.2 |
GOLGA8K
AC139426.2
|
golgin A8 family, member K Uncharacterized protein; cDNA FLJ52611 |
| chr8_+_70404996 | 0.12 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr14_+_105047478 | 0.12 |
ENST00000410013.1
|
C14orf180
|
chromosome 14 open reading frame 180 |
| chr13_-_62001982 | 0.12 |
ENST00000409186.1
|
PCDH20
|
protocadherin 20 |
| chr11_+_92085707 | 0.12 |
ENST00000525166.1
|
FAT3
|
FAT atypical cadherin 3 |
| chr1_-_226926864 | 0.12 |
ENST00000429204.1
ENST00000366784.1 |
ITPKB
|
inositol-trisphosphate 3-kinase B |
| chr1_+_46640750 | 0.12 |
ENST00000372003.1
|
TSPAN1
|
tetraspanin 1 |
| chr7_+_79998864 | 0.12 |
ENST00000435819.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr8_-_131399110 | 0.12 |
ENST00000521426.1
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr2_-_157198860 | 0.12 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr11_+_36616044 | 0.12 |
ENST00000334307.5
ENST00000531554.1 ENST00000347206.4 ENST00000534635.1 ENST00000446510.2 ENST00000530697.1 ENST00000527108.1 |
C11orf74
|
chromosome 11 open reading frame 74 |
| chr3_+_69134124 | 0.11 |
ENST00000478935.1
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr6_+_152011628 | 0.11 |
ENST00000404742.1
ENST00000440973.1 |
ESR1
|
estrogen receptor 1 |
| chr19_-_2427863 | 0.11 |
ENST00000215570.3
|
TIMM13
|
translocase of inner mitochondrial membrane 13 homolog (yeast) |
| chr15_+_30375158 | 0.11 |
ENST00000341650.6
ENST00000567927.1 |
GOLGA8J
|
golgin A8 family, member J |
| chr10_-_74385811 | 0.11 |
ENST00000603011.1
ENST00000401998.3 ENST00000361114.5 ENST00000604238.1 |
MICU1
|
mitochondrial calcium uptake 1 |
| chr4_+_26585686 | 0.11 |
ENST00000505206.1
ENST00000511789.1 |
TBC1D19
|
TBC1 domain family, member 19 |
| chr14_-_30396802 | 0.11 |
ENST00000415220.2
|
PRKD1
|
protein kinase D1 |
| chr8_-_124553437 | 0.11 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr8_-_95274536 | 0.11 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr9_+_27109392 | 0.11 |
ENST00000406359.4
|
TEK
|
TEK tyrosine kinase, endothelial |
| chr6_-_159466042 | 0.11 |
ENST00000338313.5
|
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chr21_+_17553910 | 0.11 |
ENST00000428669.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr19_+_35939154 | 0.11 |
ENST00000599180.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr20_+_43990576 | 0.11 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr13_+_73629107 | 0.11 |
ENST00000539231.1
|
KLF5
|
Kruppel-like factor 5 (intestinal) |
| chr1_-_154164534 | 0.11 |
ENST00000271850.7
ENST00000368530.2 |
TPM3
|
tropomyosin 3 |
| chr8_+_38831683 | 0.11 |
ENST00000302495.4
|
HTRA4
|
HtrA serine peptidase 4 |
| chr6_-_43595039 | 0.11 |
ENST00000307114.7
|
GTPBP2
|
GTP binding protein 2 |
| chr10_-_65028817 | 0.11 |
ENST00000542921.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr6_-_119670919 | 0.11 |
ENST00000368468.3
|
MAN1A1
|
mannosidase, alpha, class 1A, member 1 |
| chr4_+_159131346 | 0.11 |
ENST00000508243.1
ENST00000296529.6 |
TMEM144
|
transmembrane protein 144 |
| chr1_+_92632542 | 0.10 |
ENST00000409154.4
ENST00000370378.4 |
KIAA1107
|
KIAA1107 |
| chr12_+_11802753 | 0.10 |
ENST00000396373.4
|
ETV6
|
ets variant 6 |
| chr17_-_39780819 | 0.10 |
ENST00000311208.8
|
KRT17
|
keratin 17 |
| chr15_+_84841242 | 0.10 |
ENST00000558195.1
|
RP11-182J1.16
|
ubiquitin-conjugating enzyme E2Q family member 2-like |
| chr16_-_21436459 | 0.10 |
ENST00000448012.2
ENST00000504841.2 ENST00000419180.2 |
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr8_+_81397876 | 0.10 |
ENST00000430430.1
|
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr7_+_136553824 | 0.10 |
ENST00000320658.5
ENST00000453373.1 ENST00000397608.3 ENST00000402486.3 ENST00000401861.1 |
CHRM2
|
cholinergic receptor, muscarinic 2 |
| chr10_-_65028938 | 0.10 |
ENST00000402544.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr6_-_11232891 | 0.10 |
ENST00000379433.5
ENST00000379446.5 |
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr12_-_91573316 | 0.09 |
ENST00000393155.1
|
DCN
|
decorin |
| chr1_-_203320617 | 0.09 |
ENST00000354955.4
|
FMOD
|
fibromodulin |
| chr10_-_1246317 | 0.09 |
ENST00000381305.1
|
ADARB2
|
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr12_-_123921256 | 0.09 |
ENST00000280571.8
|
RILPL2
|
Rab interacting lysosomal protein-like 2 |
| chr11_-_14358620 | 0.09 |
ENST00000531421.1
|
RRAS2
|
related RAS viral (r-ras) oncogene homolog 2 |
| chr1_+_162602244 | 0.09 |
ENST00000367922.3
ENST00000367921.3 |
DDR2
|
discoidin domain receptor tyrosine kinase 2 |
| chr1_+_179712298 | 0.09 |
ENST00000341785.4
|
FAM163A
|
family with sequence similarity 163, member A |
| chr1_+_151682909 | 0.09 |
ENST00000326413.3
ENST00000442233.2 |
RIIAD1
AL589765.1
|
regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1 Uncharacterized protein; cDNA FLJ36032 fis, clone TESTI2017069 |
| chr5_+_179125368 | 0.09 |
ENST00000502296.1
ENST00000504734.1 ENST00000415618.2 |
CANX
|
calnexin |
| chr7_+_1126461 | 0.09 |
ENST00000297469.3
|
GPER1
|
G protein-coupled estrogen receptor 1 |
| chr11_+_118754475 | 0.09 |
ENST00000292174.4
|
CXCR5
|
chemokine (C-X-C motif) receptor 5 |
| chr2_+_1417228 | 0.09 |
ENST00000382269.3
ENST00000337415.3 ENST00000345913.4 ENST00000346956.3 ENST00000349624.3 ENST00000539820.1 ENST00000329066.4 ENST00000382201.3 |
TPO
|
thyroid peroxidase |
| chr7_+_1126437 | 0.09 |
ENST00000413368.1
ENST00000397092.1 |
GPER1
|
G protein-coupled estrogen receptor 1 |
| chr11_+_11863579 | 0.09 |
ENST00000399455.2
|
USP47
|
ubiquitin specific peptidase 47 |
| chr11_-_110167352 | 0.09 |
ENST00000533991.1
ENST00000528498.1 ENST00000405097.1 ENST00000528900.1 ENST00000530301.1 ENST00000343115.4 |
RDX
|
radixin |
| chr11_+_60970852 | 0.08 |
ENST00000325558.6
|
PGA3
|
pepsinogen 3, group I (pepsinogen A) |
| chr1_-_94703118 | 0.08 |
ENST00000260526.6
ENST00000370217.3 |
ARHGAP29
|
Rho GTPase activating protein 29 |
| chr2_-_97535708 | 0.08 |
ENST00000305476.5
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr16_+_15528332 | 0.08 |
ENST00000566490.1
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr1_+_150254936 | 0.08 |
ENST00000447007.1
ENST00000369095.1 ENST00000369094.1 |
C1orf51
|
chromosome 1 open reading frame 51 |
| chr14_-_107013465 | 0.08 |
ENST00000390625.2
|
IGHV3-49
|
immunoglobulin heavy variable 3-49 |
| chr1_-_77685084 | 0.08 |
ENST00000370812.3
ENST00000359130.1 ENST00000445065.1 ENST00000370813.5 |
PIGK
|
phosphatidylinositol glycan anchor biosynthesis, class K |
| chr5_-_158757895 | 0.08 |
ENST00000231228.2
|
IL12B
|
interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) |
| chr9_-_124922021 | 0.08 |
ENST00000537618.1
ENST00000373768.3 |
NDUFA8
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8, 19kDa |
| chrX_-_39186610 | 0.08 |
ENST00000429281.1
ENST00000448597.1 |
RP11-265P11.2
|
RP11-265P11.2 |
| chr15_+_41099788 | 0.08 |
ENST00000299173.10
ENST00000566407.1 |
ZFYVE19
|
zinc finger, FYVE domain containing 19 |
| chr13_-_76056250 | 0.08 |
ENST00000377636.3
ENST00000431480.2 ENST00000377625.2 ENST00000425511.1 |
TBC1D4
|
TBC1 domain family, member 4 |
| chr3_+_157261116 | 0.08 |
ENST00000468043.1
ENST00000459838.1 ENST00000461040.1 ENST00000449199.2 ENST00000426338.2 |
C3orf55
|
chromosome 3 open reading frame 55 |
| chr16_-_29415350 | 0.08 |
ENST00000524087.1
|
NPIPB11
|
nuclear pore complex interacting protein family, member B11 |
| chrX_-_119763835 | 0.08 |
ENST00000371313.2
ENST00000304661.5 |
C1GALT1C1
|
C1GALT1-specific chaperone 1 |
| chr1_+_175036966 | 0.08 |
ENST00000239462.4
|
TNN
|
tenascin N |
| chr1_+_86046433 | 0.08 |
ENST00000451137.2
|
CYR61
|
cysteine-rich, angiogenic inducer, 61 |
| chr12_+_109915179 | 0.08 |
ENST00000434735.2
|
UBE3B
|
ubiquitin protein ligase E3B |
| chr1_-_244006528 | 0.08 |
ENST00000336199.5
ENST00000263826.5 |
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr15_+_23255242 | 0.08 |
ENST00000450802.3
|
GOLGA8I
|
golgin A8 family, member I |
| chr14_+_81421861 | 0.08 |
ENST00000298171.2
|
TSHR
|
thyroid stimulating hormone receptor |
| chr8_-_70016408 | 0.07 |
ENST00000518540.1
|
RP11-600K15.1
|
RP11-600K15.1 |
| chr1_+_168545711 | 0.07 |
ENST00000367818.3
|
XCL1
|
chemokine (C motif) ligand 1 |
| chr8_-_141774467 | 0.07 |
ENST00000520151.1
ENST00000519024.1 ENST00000519465.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr1_-_156051789 | 0.07 |
ENST00000532414.2
|
MEX3A
|
mex-3 RNA binding family member A |
| chr15_+_58702742 | 0.07 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr5_-_94417339 | 0.07 |
ENST00000429576.2
ENST00000508509.1 ENST00000510732.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr18_-_52989525 | 0.07 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr15_-_59981479 | 0.07 |
ENST00000607373.1
|
BNIP2
|
BCL2/adenovirus E1B 19kDa interacting protein 2 |
| chr15_-_43785303 | 0.07 |
ENST00000382039.3
ENST00000450115.2 ENST00000382044.4 |
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr15_-_30685752 | 0.07 |
ENST00000299847.2
ENST00000397827.3 |
CHRFAM7A
|
CHRNA7 (cholinergic receptor, nicotinic, alpha 7, exons 5-10) and FAM7A (family with sequence similarity 7A, exons A-E) fusion |
| chr4_+_86749045 | 0.07 |
ENST00000514229.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chrX_+_154254960 | 0.07 |
ENST00000369498.3
|
FUNDC2
|
FUN14 domain containing 2 |
| chr13_-_67804445 | 0.07 |
ENST00000456367.1
ENST00000377861.3 ENST00000544246.1 |
PCDH9
|
protocadherin 9 |
| chr12_-_59314246 | 0.07 |
ENST00000320743.3
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3 |
| chr9_-_73483958 | 0.07 |
ENST00000377101.1
ENST00000377106.1 ENST00000360823.2 ENST00000377105.1 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr11_-_113577014 | 0.07 |
ENST00000544634.1
ENST00000539732.1 ENST00000538770.1 ENST00000536856.1 ENST00000544476.1 |
TMPRSS5
|
transmembrane protease, serine 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFATC1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 1.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.3 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.1 | 0.5 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 0.2 | GO:0030264 | nuclear fragmentation involved in apoptotic nuclear change(GO:0030264) |
| 0.1 | 1.5 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 0.2 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.3 | GO:1901910 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.6 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.3 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.1 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.2 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 1.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:2000566 | positive regulation of thymocyte migration(GO:2000412) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.0 | 0.2 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.3 | GO:0051956 | negative regulation of mitochondrial membrane potential(GO:0010917) negative regulation of amino acid transport(GO:0051956) |
| 0.0 | 0.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:2000330 | positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.2 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.0 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.5 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.1 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.0 | GO:0050720 | interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.0 | 0.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 1.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.0 | 0.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.4 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.2 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.1 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.1 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.0 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.0 | GO:0090291 | negative regulation of mesonephros development(GO:0061218) negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) negative regulation of osteoclast proliferation(GO:0090291) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 0.0 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.2 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.0 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.0 | 0.0 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.0 | 0.1 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.1 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.2 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 0.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.2 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.4 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.8 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.1 | 0.2 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.1 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.2 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.2 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.6 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.3 | GO:0052846 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.2 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.3 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0001093 | TFIIB-class transcription factor binding(GO:0001093) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.0 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.0 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |