Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
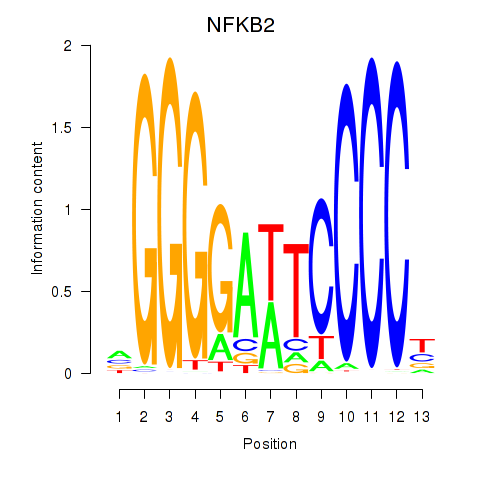
Results for NFKB2
Z-value: 2.07
Transcription factors associated with NFKB2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFKB2
|
ENSG00000077150.13 | nuclear factor kappa B subunit 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFKB2 | hg19_v2_chr10_+_104154229_104154354 | 0.90 | 1.5e-09 | Click! |
Activity profile of NFKB2 motif
Sorted Z-values of NFKB2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_123691047 | 14.79 |
ENST00000373887.3
|
TRAF1
|
TNF receptor-associated factor 1 |
| chr4_+_74702214 | 11.46 |
ENST00000226317.5
ENST00000515050.1 |
CXCL6
|
chemokine (C-X-C motif) ligand 6 |
| chr17_-_34207295 | 10.42 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr1_-_209824643 | 10.07 |
ENST00000391911.1
ENST00000415782.1 |
LAMB3
|
laminin, beta 3 |
| chr4_-_74864386 | 9.89 |
ENST00000296027.4
|
CXCL5
|
chemokine (C-X-C motif) ligand 5 |
| chr3_+_53195136 | 9.76 |
ENST00000394729.2
ENST00000330452.3 |
PRKCD
|
protein kinase C, delta |
| chr19_+_45504688 | 9.65 |
ENST00000221452.8
ENST00000540120.1 ENST00000505236.1 |
RELB
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr10_+_104154229 | 9.61 |
ENST00000428099.1
ENST00000369966.3 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr8_+_90770008 | 8.12 |
ENST00000540020.1
|
RIPK2
|
receptor-interacting serine-threonine kinase 2 |
| chr19_-_50400212 | 7.94 |
ENST00000391826.2
|
IL4I1
|
interleukin 4 induced 1 |
| chr5_-_150466692 | 7.70 |
ENST00000315050.7
ENST00000523338.1 ENST00000522100.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr8_+_90769967 | 7.16 |
ENST00000220751.4
|
RIPK2
|
receptor-interacting serine-threonine kinase 2 |
| chr6_+_14117872 | 7.12 |
ENST00000379153.3
|
CD83
|
CD83 molecule |
| chr14_-_24616426 | 7.06 |
ENST00000216802.5
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr6_-_143266297 | 6.91 |
ENST00000367603.2
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr16_+_50776021 | 6.63 |
ENST00000566679.2
ENST00000564634.1 ENST00000398568.2 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr14_-_24615805 | 6.31 |
ENST00000560410.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr9_+_115913222 | 5.68 |
ENST00000259392.3
|
SLC31A2
|
solute carrier family 31 (copper transporter), member 2 |
| chr11_-_57335750 | 5.50 |
ENST00000340573.4
|
UBE2L6
|
ubiquitin-conjugating enzyme E2L 6 |
| chr1_-_41328018 | 5.26 |
ENST00000372638.2
|
CITED4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chrX_+_68048803 | 5.26 |
ENST00000204961.4
|
EFNB1
|
ephrin-B1 |
| chr5_-_179780312 | 5.24 |
ENST00000253778.8
|
GFPT2
|
glutamine-fructose-6-phosphate transaminase 2 |
| chr1_-_8000872 | 5.23 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr21_+_27011899 | 5.01 |
ENST00000425221.2
|
JAM2
|
junctional adhesion molecule 2 |
| chr17_+_40440481 | 4.83 |
ENST00000590726.2
ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A
|
signal transducer and activator of transcription 5A |
| chr8_+_54793454 | 4.78 |
ENST00000276500.4
|
RGS20
|
regulator of G-protein signaling 20 |
| chr8_+_54793425 | 4.73 |
ENST00000522225.1
|
RGS20
|
regulator of G-protein signaling 20 |
| chr19_-_47734448 | 4.68 |
ENST00000439096.2
|
BBC3
|
BCL2 binding component 3 |
| chr19_-_4338783 | 4.51 |
ENST00000601482.1
ENST00000600324.1 |
STAP2
|
signal transducing adaptor family member 2 |
| chr4_-_146859623 | 4.50 |
ENST00000379448.4
ENST00000513320.1 |
ZNF827
|
zinc finger protein 827 |
| chr19_-_4338838 | 4.50 |
ENST00000594605.1
|
STAP2
|
signal transducing adaptor family member 2 |
| chr16_+_50775971 | 4.24 |
ENST00000311559.9
ENST00000564326.1 ENST00000566206.1 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr16_+_50775948 | 4.21 |
ENST00000569681.1
ENST00000569418.1 ENST00000540145.1 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr14_-_24615523 | 4.16 |
ENST00000559056.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr6_-_31324943 | 4.15 |
ENST00000412585.2
ENST00000434333.1 |
HLA-B
|
major histocompatibility complex, class I, B |
| chr5_-_95297534 | 4.14 |
ENST00000513343.1
ENST00000431061.2 |
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr4_+_74735102 | 4.05 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr1_+_110453109 | 4.05 |
ENST00000525659.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr6_+_18155560 | 4.04 |
ENST00000546309.2
ENST00000388870.2 ENST00000397244.1 |
KDM1B
|
lysine (K)-specific demethylase 1B |
| chr5_-_95297678 | 3.97 |
ENST00000237853.4
|
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr12_-_49259643 | 3.90 |
ENST00000309739.5
|
RND1
|
Rho family GTPase 1 |
| chr19_+_48824711 | 3.89 |
ENST00000599704.1
|
EMP3
|
epithelial membrane protein 3 |
| chr1_+_110453203 | 3.85 |
ENST00000357302.4
ENST00000344188.5 ENST00000329608.6 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr17_-_53499310 | 3.80 |
ENST00000262065.3
|
MMD
|
monocyte to macrophage differentiation-associated |
| chr12_-_57504069 | 3.73 |
ENST00000543873.2
ENST00000554663.1 ENST00000557635.1 |
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr6_+_18155632 | 3.70 |
ENST00000297792.5
|
KDM1B
|
lysine (K)-specific demethylase 1B |
| chr21_+_27011584 | 3.63 |
ENST00000400532.1
ENST00000480456.1 ENST00000312957.5 |
JAM2
|
junctional adhesion molecule 2 |
| chr10_+_13142075 | 3.61 |
ENST00000378757.2
ENST00000430081.1 ENST00000378752.3 ENST00000378748.3 |
OPTN
|
optineurin |
| chr16_+_57653989 | 3.51 |
ENST00000567835.1
ENST00000569372.1 ENST00000563548.1 ENST00000562003.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr10_+_13141585 | 3.50 |
ENST00000378764.2
|
OPTN
|
optineurin |
| chr10_+_13142225 | 3.47 |
ENST00000378747.3
|
OPTN
|
optineurin |
| chr16_+_57653854 | 3.44 |
ENST00000568908.1
ENST00000568909.1 ENST00000566778.1 ENST00000561988.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr1_+_65613217 | 3.42 |
ENST00000545314.1
|
AK4
|
adenylate kinase 4 |
| chr4_-_74904398 | 3.37 |
ENST00000296026.4
|
CXCL3
|
chemokine (C-X-C motif) ligand 3 |
| chr2_-_136743169 | 3.35 |
ENST00000264161.4
|
DARS
|
aspartyl-tRNA synthetase |
| chr20_-_22559211 | 3.28 |
ENST00000564492.1
|
LINC00261
|
long intergenic non-protein coding RNA 261 |
| chr1_-_47082495 | 3.18 |
ENST00000545730.1
ENST00000531769.1 ENST00000319928.3 |
MKNK1
MOB3C
|
MAP kinase interacting serine/threonine kinase 1 MOB kinase activator 3C |
| chr8_-_145582118 | 2.93 |
ENST00000455319.2
ENST00000331890.5 |
FBXL6
|
F-box and leucine-rich repeat protein 6 |
| chr1_+_110453462 | 2.80 |
ENST00000488198.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr14_+_24605389 | 2.79 |
ENST00000382708.3
ENST00000561435.1 |
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr19_+_45147313 | 2.77 |
ENST00000406449.4
|
PVR
|
poliovirus receptor |
| chr19_+_45147098 | 2.77 |
ENST00000425690.3
ENST00000344956.4 ENST00000403059.4 |
PVR
|
poliovirus receptor |
| chr8_+_145582231 | 2.71 |
ENST00000526338.1
ENST00000402965.1 ENST00000534725.1 ENST00000532887.1 ENST00000329994.2 |
SLC52A2
|
solute carrier family 52 (riboflavin transporter), member 2 |
| chr20_-_48330377 | 2.59 |
ENST00000371711.4
|
B4GALT5
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
| chr5_+_60241020 | 2.59 |
ENST00000511107.1
ENST00000502658.1 |
NDUFAF2
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 2 |
| chr10_+_73724123 | 2.58 |
ENST00000373115.4
|
CHST3
|
carbohydrate (chondroitin 6) sulfotransferase 3 |
| chr16_+_4674814 | 2.58 |
ENST00000415496.1
ENST00000587747.1 ENST00000399577.5 ENST00000588994.1 ENST00000586183.1 |
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr15_+_63414760 | 2.56 |
ENST00000557972.1
|
LACTB
|
lactamase, beta |
| chr3_+_9691117 | 2.56 |
ENST00000353332.5
ENST00000420925.1 ENST00000296003.4 ENST00000351233.5 |
MTMR14
|
myotubularin related protein 14 |
| chr19_+_13135790 | 2.55 |
ENST00000358552.3
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr12_-_77272765 | 2.54 |
ENST00000547435.1
ENST00000552330.1 ENST00000546966.1 ENST00000311083.5 |
CSRP2
|
cysteine and glycine-rich protein 2 |
| chr1_+_170633047 | 2.53 |
ENST00000239461.6
ENST00000497230.2 |
PRRX1
|
paired related homeobox 1 |
| chr10_-_86001210 | 2.50 |
ENST00000372105.3
|
LRIT1
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1 |
| chr11_+_35639735 | 2.48 |
ENST00000317811.4
|
FJX1
|
four jointed box 1 (Drosophila) |
| chr2_-_136743039 | 2.43 |
ENST00000537273.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr12_-_322504 | 2.38 |
ENST00000424061.2
|
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr5_+_176730769 | 2.36 |
ENST00000303204.4
ENST00000503216.1 |
PRELID1
|
PRELI domain containing 1 |
| chr16_-_81110683 | 2.28 |
ENST00000565253.1
ENST00000378611.4 ENST00000299578.5 |
C16orf46
|
chromosome 16 open reading frame 46 |
| chr5_+_60240943 | 2.26 |
ENST00000296597.5
|
NDUFAF2
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 2 |
| chr20_-_48532019 | 2.21 |
ENST00000289431.5
|
SPATA2
|
spermatogenesis associated 2 |
| chr16_-_57318566 | 2.17 |
ENST00000569059.1
ENST00000219207.5 |
PLLP
|
plasmolipin |
| chrX_+_150151824 | 2.17 |
ENST00000455596.1
ENST00000448905.2 |
HMGB3
|
high mobility group box 3 |
| chr6_+_80341000 | 2.13 |
ENST00000369838.4
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2 |
| chr7_+_100547156 | 2.13 |
ENST00000379458.4
|
MUC3A
|
Protein LOC100131514 |
| chr14_+_24605361 | 2.12 |
ENST00000206451.6
ENST00000559123.1 |
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr8_+_145582217 | 2.12 |
ENST00000530047.1
ENST00000527078.1 |
SLC52A2
|
solute carrier family 52 (riboflavin transporter), member 2 |
| chr3_+_111717600 | 2.11 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr16_-_4466622 | 2.11 |
ENST00000570645.1
ENST00000574025.1 ENST00000572898.1 ENST00000537233.2 ENST00000571059.1 ENST00000251166.4 |
CORO7
|
coronin 7 |
| chr3_+_182971583 | 2.05 |
ENST00000460419.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr1_+_2487800 | 2.00 |
ENST00000355716.4
|
TNFRSF14
|
tumor necrosis factor receptor superfamily, member 14 |
| chr3_+_182971018 | 1.99 |
ENST00000326505.3
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr6_+_44095347 | 1.98 |
ENST00000323267.6
|
TMEM63B
|
transmembrane protein 63B |
| chr4_-_146859787 | 1.97 |
ENST00000508784.1
|
ZNF827
|
zinc finger protein 827 |
| chr13_-_73356009 | 1.96 |
ENST00000377780.4
ENST00000377767.4 |
DIS3
|
DIS3 mitotic control homolog (S. cerevisiae) |
| chr22_-_19512893 | 1.96 |
ENST00000403084.1
ENST00000413119.2 |
CLDN5
|
claudin 5 |
| chr17_-_1389419 | 1.96 |
ENST00000575158.1
|
MYO1C
|
myosin IC |
| chrX_-_118986911 | 1.91 |
ENST00000276201.2
ENST00000345865.2 |
UPF3B
|
UPF3 regulator of nonsense transcripts homolog B (yeast) |
| chr11_+_6625046 | 1.91 |
ENST00000396751.2
|
ILK
|
integrin-linked kinase |
| chr5_-_146889619 | 1.90 |
ENST00000343218.5
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr11_-_104840093 | 1.89 |
ENST00000417440.2
ENST00000444739.2 |
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr11_-_75062829 | 1.87 |
ENST00000393505.4
|
ARRB1
|
arrestin, beta 1 |
| chr4_-_74964904 | 1.86 |
ENST00000508487.2
|
CXCL2
|
chemokine (C-X-C motif) ligand 2 |
| chr11_+_6624955 | 1.84 |
ENST00000299421.4
ENST00000537806.1 |
ILK
|
integrin-linked kinase |
| chr3_+_111718036 | 1.82 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr17_+_21191341 | 1.80 |
ENST00000526076.2
ENST00000361818.5 ENST00000316920.6 |
MAP2K3
|
mitogen-activated protein kinase kinase 3 |
| chr5_+_170846640 | 1.79 |
ENST00000274625.5
|
FGF18
|
fibroblast growth factor 18 |
| chr19_+_45251804 | 1.79 |
ENST00000164227.5
|
BCL3
|
B-cell CLL/lymphoma 3 |
| chr4_-_5890145 | 1.79 |
ENST00000397890.2
|
CRMP1
|
collapsin response mediator protein 1 |
| chr14_-_51297197 | 1.78 |
ENST00000382043.4
|
NIN
|
ninein (GSK3B interacting protein) |
| chr17_-_1389228 | 1.78 |
ENST00000438665.2
|
MYO1C
|
myosin IC |
| chr5_+_176731572 | 1.75 |
ENST00000503853.1
|
PRELID1
|
PRELI domain containing 1 |
| chr19_-_55677920 | 1.75 |
ENST00000524407.2
ENST00000526003.1 ENST00000534170.1 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr1_-_155232047 | 1.72 |
ENST00000302631.3
|
SCAMP3
|
secretory carrier membrane protein 3 |
| chr1_-_151299842 | 1.65 |
ENST00000438243.2
ENST00000489223.2 ENST00000368873.1 ENST00000430800.1 ENST00000368872.1 |
PI4KB
|
phosphatidylinositol 4-kinase, catalytic, beta |
| chr2_+_190306159 | 1.64 |
ENST00000314761.4
|
WDR75
|
WD repeat domain 75 |
| chr11_-_75062730 | 1.63 |
ENST00000420843.2
ENST00000360025.3 |
ARRB1
|
arrestin, beta 1 |
| chr3_-_182698381 | 1.63 |
ENST00000292782.4
|
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr7_-_98467543 | 1.63 |
ENST00000345589.4
|
TMEM130
|
transmembrane protein 130 |
| chr16_+_70613770 | 1.61 |
ENST00000429149.2
ENST00000563721.2 |
IL34
|
interleukin 34 |
| chr11_-_76381781 | 1.61 |
ENST00000260061.5
ENST00000404995.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr7_-_98467489 | 1.60 |
ENST00000416379.2
|
TMEM130
|
transmembrane protein 130 |
| chr7_-_98467629 | 1.59 |
ENST00000339375.4
|
TMEM130
|
transmembrane protein 130 |
| chr2_+_64681641 | 1.58 |
ENST00000409537.2
|
LGALSL
|
lectin, galactoside-binding-like |
| chr11_-_3862206 | 1.56 |
ENST00000351018.4
|
RHOG
|
ras homolog family member G |
| chr1_+_183441618 | 1.56 |
ENST00000507691.2
ENST00000508461.1 ENST00000419169.1 ENST00000347615.2 ENST00000507469.1 ENST00000515829.2 |
SMG7
|
SMG7 nonsense mediated mRNA decay factor |
| chr10_-_36813162 | 1.55 |
ENST00000440465.1
|
NAMPTL
|
nicotinamide phosphoribosyltransferase-like |
| chr16_-_66959429 | 1.55 |
ENST00000420652.1
ENST00000299759.6 |
RRAD
|
Ras-related associated with diabetes |
| chr16_-_4465886 | 1.53 |
ENST00000539968.1
|
CORO7
|
coronin 7 |
| chr15_+_92396920 | 1.53 |
ENST00000318445.6
|
SLCO3A1
|
solute carrier organic anion transporter family, member 3A1 |
| chr20_+_35090150 | 1.52 |
ENST00000340491.4
|
DLGAP4
|
discs, large (Drosophila) homolog-associated protein 4 |
| chr19_-_55677999 | 1.52 |
ENST00000532817.1
ENST00000527223.2 ENST00000391720.4 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr17_+_76164639 | 1.48 |
ENST00000225777.3
ENST00000585591.1 ENST00000589711.1 ENST00000588282.1 ENST00000589168.1 |
SYNGR2
|
synaptogyrin 2 |
| chr22_-_21984282 | 1.48 |
ENST00000398873.3
ENST00000292778.6 |
YDJC
|
YdjC homolog (bacterial) |
| chr5_-_176730676 | 1.48 |
ENST00000393611.2
ENST00000303251.6 ENST00000303270.6 |
RAB24
|
RAB24, member RAS oncogene family |
| chrX_-_23761317 | 1.47 |
ENST00000492081.1
ENST00000379303.5 ENST00000336430.7 |
ACOT9
|
acyl-CoA thioesterase 9 |
| chr10_+_118350522 | 1.47 |
ENST00000530319.1
ENST00000527980.1 ENST00000471549.1 ENST00000534537.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr10_+_118350468 | 1.45 |
ENST00000358834.4
ENST00000528052.1 ENST00000442761.1 |
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr11_+_46639071 | 1.44 |
ENST00000580238.1
ENST00000581416.1 ENST00000529655.1 ENST00000533325.1 ENST00000581438.1 ENST00000583249.1 ENST00000530500.1 ENST00000526508.1 ENST00000578626.1 ENST00000577256.1 ENST00000524625.1 ENST00000582547.1 ENST00000359513.4 ENST00000528494.1 |
ATG13
|
autophagy related 13 |
| chr1_+_183441600 | 1.44 |
ENST00000367537.3
|
SMG7
|
SMG7 nonsense mediated mRNA decay factor |
| chr8_+_32406179 | 1.40 |
ENST00000405005.3
|
NRG1
|
neuregulin 1 |
| chr16_-_74734742 | 1.39 |
ENST00000308807.7
ENST00000573267.1 |
MLKL
|
mixed lineage kinase domain-like |
| chr14_-_51297837 | 1.36 |
ENST00000245441.5
ENST00000389868.3 ENST00000382041.3 ENST00000324330.9 ENST00000453196.1 ENST00000453401.2 |
NIN
|
ninein (GSK3B interacting protein) |
| chr1_-_89458287 | 1.36 |
ENST00000370485.2
|
CCBL2
|
cysteine conjugate-beta lyase 2 |
| chrX_-_15353629 | 1.36 |
ENST00000333590.4
ENST00000428964.1 ENST00000542278.1 |
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A |
| chr20_-_62258394 | 1.35 |
ENST00000370077.1
|
GMEB2
|
glucocorticoid modulatory element binding protein 2 |
| chr17_+_76165213 | 1.32 |
ENST00000590201.1
|
SYNGR2
|
synaptogyrin 2 |
| chr16_-_1429674 | 1.32 |
ENST00000403703.1
ENST00000397464.1 ENST00000402641.2 |
UNKL
|
unkempt family zinc finger-like |
| chr1_-_89458415 | 1.31 |
ENST00000321792.5
ENST00000370491.3 |
RBMXL1
CCBL2
|
RNA binding motif protein, X-linked-like 1 cysteine conjugate-beta lyase 2 |
| chr19_-_10450328 | 1.30 |
ENST00000160262.5
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr8_+_38677850 | 1.29 |
ENST00000518809.1
ENST00000520611.1 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr4_-_169239921 | 1.29 |
ENST00000514995.1
ENST00000393743.3 |
DDX60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chrX_+_64887512 | 1.28 |
ENST00000360270.5
|
MSN
|
moesin |
| chr17_-_42908155 | 1.26 |
ENST00000426548.1
ENST00000590758.1 ENST00000591424.1 |
GJC1
|
gap junction protein, gamma 1, 45kDa |
| chr11_+_58910295 | 1.26 |
ENST00000420244.1
|
FAM111A
|
family with sequence similarity 111, member A |
| chr5_+_67511524 | 1.26 |
ENST00000521381.1
ENST00000521657.1 |
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr15_+_92937144 | 1.26 |
ENST00000539113.1
ENST00000555434.1 |
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr11_+_706113 | 1.26 |
ENST00000318562.8
ENST00000533256.1 ENST00000534755.1 |
EPS8L2
|
EPS8-like 2 |
| chr6_+_135502408 | 1.26 |
ENST00000341911.5
ENST00000442647.2 ENST00000316528.8 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr17_-_3819751 | 1.25 |
ENST00000225538.3
|
P2RX1
|
purinergic receptor P2X, ligand-gated ion channel, 1 |
| chr18_+_74240610 | 1.24 |
ENST00000578092.1
ENST00000578613.1 ENST00000583578.1 |
LINC00908
|
long intergenic non-protein coding RNA 908 |
| chr1_+_90287480 | 1.22 |
ENST00000394593.3
|
LRRC8D
|
leucine rich repeat containing 8 family, member D |
| chr1_+_19578033 | 1.20 |
ENST00000330263.4
|
MRTO4
|
mRNA turnover 4 homolog (S. cerevisiae) |
| chr17_-_1395954 | 1.20 |
ENST00000359786.5
|
MYO1C
|
myosin IC |
| chr3_+_52232102 | 1.17 |
ENST00000469224.1
ENST00000394965.2 ENST00000310271.2 ENST00000484952.1 |
ALAS1
|
aminolevulinate, delta-, synthase 1 |
| chr19_+_33072373 | 1.17 |
ENST00000586035.1
|
PDCD5
|
programmed cell death 5 |
| chr16_+_31119615 | 1.16 |
ENST00000394950.3
ENST00000287507.3 ENST00000219794.6 ENST00000561755.1 |
BCKDK
|
branched chain ketoacid dehydrogenase kinase |
| chr9_+_17579084 | 1.15 |
ENST00000380607.4
|
SH3GL2
|
SH3-domain GRB2-like 2 |
| chr12_-_50294033 | 1.14 |
ENST00000552669.1
|
FAIM2
|
Fas apoptotic inhibitory molecule 2 |
| chr11_-_46142505 | 1.13 |
ENST00000524497.1
ENST00000418153.2 |
PHF21A
|
PHD finger protein 21A |
| chr17_+_8924837 | 1.13 |
ENST00000173229.2
|
NTN1
|
netrin 1 |
| chr15_-_72767490 | 1.13 |
ENST00000565181.1
|
RP11-1007O24.3
|
RP11-1007O24.3 |
| chr3_-_50329990 | 1.12 |
ENST00000417626.2
|
IFRD2
|
interferon-related developmental regulator 2 |
| chr11_+_58910201 | 1.12 |
ENST00000528737.1
|
FAM111A
|
family with sequence similarity 111, member A |
| chr20_-_62462566 | 1.11 |
ENST00000245663.4
ENST00000302995.2 |
ZBTB46
|
zinc finger and BTB domain containing 46 |
| chr19_+_39390587 | 1.10 |
ENST00000572515.1
ENST00000392079.3 ENST00000575359.1 ENST00000313582.5 |
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr17_+_28884130 | 1.09 |
ENST00000580161.1
|
TBC1D29
|
TBC1 domain family, member 29 |
| chr5_+_137203465 | 1.08 |
ENST00000239926.4
|
MYOT
|
myotilin |
| chrX_-_2418936 | 1.08 |
ENST00000461691.1
ENST00000381223.4 ENST00000381222.2 ENST00000412516.2 ENST00000334651.5 |
ZBED1
DHRSX
|
zinc finger, BED-type containing 1 dehydrogenase/reductase (SDR family) X-linked |
| chr16_-_4466565 | 1.07 |
ENST00000572467.1
ENST00000423908.2 ENST00000572044.1 ENST00000571052.1 |
CORO7-PAM16
CORO7
|
CORO7-PAM16 readthrough coronin 7 |
| chr6_+_137143694 | 1.06 |
ENST00000367756.4
ENST00000541292.1 ENST00000318471.4 |
PEX7
|
peroxisomal biogenesis factor 7 |
| chr6_-_43484621 | 1.06 |
ENST00000506469.1
ENST00000503972.1 |
YIPF3
|
Yip1 domain family, member 3 |
| chr10_-_133795416 | 1.06 |
ENST00000540159.1
ENST00000368636.4 |
BNIP3
|
BCL2/adenovirus E1B 19kDa interacting protein 3 |
| chr19_+_38794797 | 1.06 |
ENST00000301246.5
ENST00000588605.1 |
C19orf33
|
chromosome 19 open reading frame 33 |
| chr19_+_45842445 | 1.06 |
ENST00000598357.1
|
L47234.1
|
Uncharacterized protein |
| chr14_+_24616588 | 1.02 |
ENST00000324103.6
ENST00000559260.1 |
RNF31
|
ring finger protein 31 |
| chr8_+_120079478 | 1.02 |
ENST00000332843.2
|
COLEC10
|
collectin sub-family member 10 (C-type lectin) |
| chr2_-_89568263 | 1.01 |
ENST00000473726.1
|
IGKV1-33
|
immunoglobulin kappa variable 1-33 |
| chr15_-_89438742 | 1.01 |
ENST00000562281.1
ENST00000562889.1 ENST00000359595.3 |
HAPLN3
|
hyaluronan and proteoglycan link protein 3 |
| chr4_-_860950 | 1.01 |
ENST00000511980.1
ENST00000510799.1 |
GAK
|
cyclin G associated kinase |
| chr3_-_50329835 | 1.01 |
ENST00000429673.2
|
IFRD2
|
interferon-related developmental regulator 2 |
| chr5_-_1445539 | 1.00 |
ENST00000270349.9
|
SLC6A3
|
solute carrier family 6 (neurotransmitter transporter), member 3 |
| chr12_-_130388211 | 0.99 |
ENST00000422113.2
|
TMEM132D
|
transmembrane protein 132D |
| chr12_+_100661156 | 0.98 |
ENST00000360820.2
|
SCYL2
|
SCY1-like 2 (S. cerevisiae) |
| chr1_-_146082633 | 0.98 |
ENST00000605317.1
ENST00000604938.1 ENST00000339388.5 |
NBPF11
|
neuroblastoma breakpoint family, member 11 |
| chr20_+_58713514 | 0.97 |
ENST00000432910.1
|
RP5-1043L13.1
|
RP5-1043L13.1 |
| chr22_-_37640456 | 0.96 |
ENST00000405484.1
ENST00000441619.1 ENST00000406508.1 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr1_-_150207017 | 0.96 |
ENST00000369119.3
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr6_-_43484718 | 0.95 |
ENST00000372422.2
|
YIPF3
|
Yip1 domain family, member 3 |
| chr2_-_27718052 | 0.95 |
ENST00000264703.3
|
FNDC4
|
fibronectin type III domain containing 4 |
| chr2_+_89952792 | 0.95 |
ENST00000390265.2
|
IGKV1D-33
|
immunoglobulin kappa variable 1D-33 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFKB2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 15.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 3.1 | 15.3 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 2.8 | 11.4 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 2.6 | 10.4 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 2.4 | 9.8 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 2.3 | 11.4 | GO:1903971 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 1.9 | 5.8 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 1.9 | 7.7 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 1.8 | 5.5 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 1.2 | 4.8 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 1.2 | 10.6 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 1.1 | 32.7 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 1.0 | 4.8 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.9 | 3.7 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.9 | 7.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.9 | 5.2 | GO:2001183 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.9 | 7.7 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.8 | 4.9 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.7 | 6.6 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.6 | 1.8 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.6 | 4.2 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.6 | 5.7 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.5 | 7.9 | GO:1902222 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.4 | 1.3 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.4 | 4.1 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.4 | 5.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.4 | 2.0 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.4 | 1.5 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.4 | 1.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.4 | 9.7 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.4 | 1.1 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.4 | 10.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 4.8 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.3 | 3.4 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.3 | 1.0 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.3 | 2.3 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.3 | 2.0 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.3 | 1.3 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.3 | 1.3 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.3 | 1.2 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.3 | 3.5 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.3 | 1.6 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.3 | 0.5 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.3 | 0.8 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.2 | 1.2 | GO:0002351 | serotonin production involved in inflammatory response(GO:0002351) serotonin secretion involved in inflammatory response(GO:0002442) serotonin secretion by platelet(GO:0002554) |
| 0.2 | 2.0 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.2 | 3.9 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.2 | 0.7 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.2 | 2.5 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.2 | 24.2 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.2 | 0.7 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.2 | 1.8 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.2 | 3.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.2 | 1.8 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 | 0.7 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.2 | 2.6 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.2 | 0.6 | GO:1900222 | regulation of cell growth by extracellular stimulus(GO:0001560) negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.2 | 0.6 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.2 | 17.6 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.2 | 1.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.2 | 1.3 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.2 | 0.7 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.2 | 1.6 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.2 | 0.7 | GO:0002265 | astrocyte activation involved in immune response(GO:0002265) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.2 | 0.5 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) regulation of AV node cell action potential(GO:0098904) |
| 0.2 | 2.2 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.2 | 0.5 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.2 | 1.1 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) negative regulation of macrophage differentiation(GO:0045650) |
| 0.2 | 0.6 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.2 | 0.9 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.4 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.9 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 | 0.9 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.1 | 3.8 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 1.1 | GO:0033564 | Cdc42 protein signal transduction(GO:0032488) anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 1.0 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 0.7 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.1 | 3.7 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 0.7 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.1 | 0.4 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 0.5 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.1 | 1.8 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.4 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.1 | 2.4 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.1 | 0.2 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.8 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.1 | 9.0 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 4.7 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 1.7 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 8.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 2.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 3.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.8 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 5.3 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.1 | 0.9 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.1 | 0.3 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.1 | 0.7 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.1 | 1.2 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.1 | 8.1 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 0.7 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 1.1 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.1 | 1.1 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 0.6 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.1 | 0.3 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.1 | 1.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 1.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 1.6 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.1 | 3.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 0.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 1.3 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.1 | 1.6 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.2 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.1 | 0.4 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 2.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.9 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 0.3 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.1 | 1.0 | GO:0042416 | dopamine biosynthetic process(GO:0042416) |
| 0.1 | 0.5 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.1 | 0.3 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 2.8 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 1.4 | GO:0098780 | response to mitochondrial depolarisation(GO:0098780) |
| 0.1 | 0.4 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.5 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.6 | GO:1903546 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 0.6 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.1 | GO:0034128 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.1 | 0.6 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 0.6 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.1 | 0.5 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.1 | 0.5 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 1.1 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.3 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.4 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.0 | 0.3 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.5 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.9 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 1.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 1.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 1.4 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.8 | GO:0008595 | blastoderm segmentation(GO:0007350) tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 3.9 | GO:0007004 | telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 0.5 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.4 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.5 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 3.5 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 1.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.5 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 1.1 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.3 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.3 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 1.6 | GO:0050710 | negative regulation of cytokine secretion(GO:0050710) |
| 0.0 | 0.5 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.7 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 4.8 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 2.4 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 1.4 | GO:0090280 | positive regulation of calcium ion import(GO:0090280) |
| 0.0 | 0.9 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 1.3 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.2 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) endothelial cell-cell adhesion(GO:0071603) |
| 0.0 | 0.8 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.7 | GO:0070266 | necroptotic process(GO:0070266) |
| 0.0 | 0.3 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.8 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.1 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 1.4 | GO:0032392 | DNA geometric change(GO:0032392) |
| 0.0 | 0.6 | GO:1901099 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.8 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.9 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.2 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 1.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.5 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 1.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 22.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 2.8 | 11.4 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 1.9 | 11.4 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 1.3 | 10.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 1.2 | 4.9 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.4 | 3.6 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.4 | 4.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.3 | 3.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.3 | 6.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.3 | 1.3 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.3 | 0.8 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.3 | 2.6 | GO:0060091 | kinocilium(GO:0060091) |
| 0.3 | 14.9 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.2 | 1.8 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.2 | 1.9 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.2 | 1.0 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.2 | 0.8 | GO:0044308 | axonal spine(GO:0044308) |
| 0.2 | 0.5 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.2 | 1.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.2 | 4.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 1.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 12.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 1.6 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 1.8 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 2.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 7.7 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 1.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.6 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 7.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 3.9 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 1.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 2.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.9 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 1.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 10.4 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 0.7 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 1.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 3.7 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.0 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.5 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 0.7 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 2.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.6 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 1.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 2.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 1.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 6.7 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 8.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 5.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.2 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.5 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 1.0 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 1.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.4 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 2.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 2.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.9 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 3.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 3.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.3 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 7.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.2 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 43.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 10.9 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 1.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 2.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 1.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 2.4 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 6.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.1 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 4.4 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.4 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 1.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.3 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 22.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 2.6 | 7.9 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 2.0 | 31.6 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 2.0 | 9.8 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 1.6 | 13.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 1.5 | 10.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 1.3 | 14.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 1.3 | 4.0 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 1.3 | 7.7 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 1.2 | 3.5 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 1.2 | 5.8 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 1.1 | 14.8 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 1.0 | 4.8 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.9 | 14.8 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.8 | 6.6 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.8 | 4.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.7 | 3.4 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.6 | 1.8 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.6 | 5.7 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.5 | 2.0 | GO:0004803 | transposase activity(GO:0004803) |
| 0.4 | 2.4 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.4 | 10.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.4 | 1.2 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.4 | 1.2 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.4 | 3.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.4 | 1.5 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.4 | 2.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.3 | 1.0 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 0.3 | 1.3 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.3 | 2.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.3 | 7.8 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.2 | 2.6 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.2 | 1.6 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 0.7 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.2 | 0.7 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.2 | 1.8 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 4.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 2.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 0.7 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.2 | 0.7 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.2 | 6.8 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 3.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.2 | 4.8 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.2 | 1.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 1.3 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.2 | 1.8 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.4 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 3.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.7 | GO:0042954 | apolipoprotein receptor activity(GO:0030226) lipoprotein transporter activity(GO:0042954) |
| 0.1 | 0.5 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 0.5 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.1 | 0.9 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 8.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 1.8 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 1.6 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 3.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 1.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 1.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 0.7 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 1.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 1.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 3.9 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 5.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.8 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 0.9 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.4 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.1 | 1.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.9 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.5 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 1.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 4.8 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.2 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.1 | 0.3 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.1 | 0.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 1.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 1.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.3 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.1 | 1.8 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 0.4 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.1 | 0.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.9 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 0.6 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.4 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 1.8 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.0 | 0.9 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.8 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.5 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 9.5 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.6 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 1.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 10.8 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 1.1 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 2.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.5 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.5 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.8 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 3.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.5 | GO:0016502 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.0 | 0.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 3.5 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.0 | 4.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 8.0 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.4 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.8 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 1.8 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.7 | 30.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.4 | 10.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.4 | 19.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.3 | 12.0 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.3 | 17.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.3 | 9.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.3 | 4.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.2 | 5.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.2 | 1.1 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.2 | 10.1 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.2 | 2.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.2 | 8.5 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 5.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 13.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 5.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 6.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 29.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 2.6 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 4.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.1 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 1.3 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 1.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.5 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.9 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 2.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 4.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 2.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 2.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 2.4 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.7 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.6 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 2.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 15.3 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.6 | 41.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.4 | 4.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.4 | 10.7 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.4 | 20.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.3 | 5.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.3 | 22.4 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.3 | 6.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.3 | 19.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.2 | 3.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 4.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 0.7 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.2 | 6.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 4.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 3.5 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.2 | 6.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 15.9 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.2 | 4.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 2.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.2 | 2.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 3.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 3.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.8 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 2.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 3.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.8 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 1.8 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 1.7 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 9.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 1.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 1.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 0.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 3.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 2.4 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 2.8 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 1.8 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 1.0 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.0 | 0.8 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 1.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.7 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.8 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 1.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 2.9 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.4 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.4 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 1.1 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.7 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |