Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
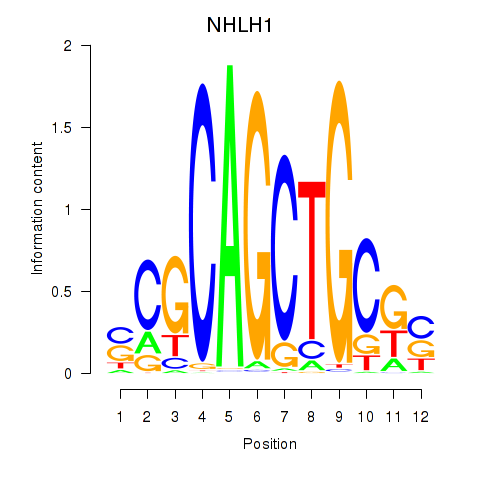
Results for NHLH1
Z-value: 0.82
Transcription factors associated with NHLH1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NHLH1
|
ENSG00000171786.5 | nescient helix-loop-helix 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NHLH1 | hg19_v2_chr1_+_160336851_160336868 | -0.03 | 8.8e-01 | Click! |
Activity profile of NHLH1 motif
Sorted Z-values of NHLH1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_39382900 | 4.03 |
ENST00000377721.3
ENST00000455970.2 |
KRTAP9-2
|
keratin associated protein 9-2 |
| chr4_-_186696425 | 1.64 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr4_-_186697044 | 1.51 |
ENST00000437304.2
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr14_+_63671577 | 1.17 |
ENST00000555125.1
|
RHOJ
|
ras homolog family member J |
| chr13_-_72440901 | 0.98 |
ENST00000359684.2
|
DACH1
|
dachshund homolog 1 (Drosophila) |
| chr5_+_56111361 | 0.96 |
ENST00000399503.3
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr11_-_10830463 | 0.92 |
ENST00000527419.1
ENST00000530211.1 ENST00000530702.1 ENST00000524932.1 ENST00000532570.1 |
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr20_+_34042962 | 0.86 |
ENST00000446710.1
ENST00000420564.1 |
CEP250
|
centrosomal protein 250kDa |
| chr11_-_1643368 | 0.84 |
ENST00000399682.1
|
KRTAP5-4
|
keratin associated protein 5-4 |
| chr14_+_63671105 | 0.76 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr18_+_77623668 | 0.76 |
ENST00000316249.3
|
KCNG2
|
potassium voltage-gated channel, subfamily G, member 2 |
| chr12_-_118406777 | 0.73 |
ENST00000339824.5
|
KSR2
|
kinase suppressor of ras 2 |
| chr6_+_35995552 | 0.72 |
ENST00000468133.1
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr7_+_150065879 | 0.70 |
ENST00000397281.2
ENST00000444957.1 ENST00000466559.1 ENST00000489432.2 ENST00000475514.1 ENST00000482680.1 ENST00000488943.1 ENST00000518514.1 ENST00000478789.1 |
REPIN1
ZNF775
|
replication initiator 1 zinc finger protein 775 |
| chr8_+_98788057 | 0.67 |
ENST00000517924.1
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta |
| chr19_+_14184370 | 0.66 |
ENST00000590772.1
|
hsa-mir-1199
|
hsa-mir-1199 |
| chr2_+_191513789 | 0.65 |
ENST00000409581.1
|
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1) |
| chr14_-_54423529 | 0.65 |
ENST00000245451.4
ENST00000559087.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr10_-_101945771 | 0.65 |
ENST00000370408.2
ENST00000407654.3 |
ERLIN1
|
ER lipid raft associated 1 |
| chr22_-_37882395 | 0.65 |
ENST00000416983.3
ENST00000424765.2 ENST00000356998.3 |
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr20_-_39317868 | 0.63 |
ENST00000373313.2
|
MAFB
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B |
| chr9_-_139581875 | 0.59 |
ENST00000371696.2
|
AGPAT2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 |
| chr3_-_32022733 | 0.59 |
ENST00000438237.2
ENST00000396556.2 |
OSBPL10
|
oxysterol binding protein-like 10 |
| chr1_-_47184745 | 0.59 |
ENST00000544071.1
|
EFCAB14
|
EF-hand calcium binding domain 14 |
| chr1_-_209979465 | 0.58 |
ENST00000542854.1
|
IRF6
|
interferon regulatory factor 6 |
| chr9_-_139581848 | 0.58 |
ENST00000538402.1
ENST00000371694.3 |
AGPAT2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 |
| chr1_+_78511586 | 0.58 |
ENST00000370759.3
|
GIPC2
|
GIPC PDZ domain containing family, member 2 |
| chr1_-_182360918 | 0.56 |
ENST00000339526.4
|
GLUL
|
glutamate-ammonia ligase |
| chr17_-_54991369 | 0.56 |
ENST00000537230.1
|
TRIM25
|
tripartite motif containing 25 |
| chrX_-_118827113 | 0.55 |
ENST00000394617.2
|
SEPT6
|
septin 6 |
| chr15_-_69113218 | 0.54 |
ENST00000560303.1
ENST00000465139.2 |
ANP32A
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr11_-_10829851 | 0.54 |
ENST00000532082.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr1_-_92351769 | 0.54 |
ENST00000212355.4
|
TGFBR3
|
transforming growth factor, beta receptor III |
| chr6_+_35995488 | 0.54 |
ENST00000229795.3
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr1_-_209979375 | 0.53 |
ENST00000367021.3
|
IRF6
|
interferon regulatory factor 6 |
| chr13_+_31480328 | 0.53 |
ENST00000380482.4
|
MEDAG
|
mesenteric estrogen-dependent adipogenesis |
| chr4_+_86748898 | 0.52 |
ENST00000509300.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chrX_-_118826784 | 0.52 |
ENST00000394616.4
|
SEPT6
|
septin 6 |
| chr16_+_69373471 | 0.51 |
ENST00000569637.2
|
NIP7
|
NIP7, nucleolar pre-rRNA processing protein |
| chrX_+_84499081 | 0.51 |
ENST00000276123.3
|
ZNF711
|
zinc finger protein 711 |
| chr4_-_80994619 | 0.50 |
ENST00000404191.1
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr1_-_21606013 | 0.50 |
ENST00000357071.4
|
ECE1
|
endothelin converting enzyme 1 |
| chr22_-_21905120 | 0.50 |
ENST00000331505.5
|
RIMBP3C
|
RIMS binding protein 3C |
| chr8_-_101963482 | 0.49 |
ENST00000419477.2
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr12_+_57482665 | 0.49 |
ENST00000300131.3
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr12_-_120554534 | 0.49 |
ENST00000538903.1
ENST00000534951.1 |
RAB35
|
RAB35, member RAS oncogene family |
| chr4_+_86749045 | 0.49 |
ENST00000514229.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr8_+_98788003 | 0.48 |
ENST00000521545.2
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta |
| chr6_-_75915757 | 0.48 |
ENST00000322507.8
|
COL12A1
|
collagen, type XII, alpha 1 |
| chr6_+_35995531 | 0.47 |
ENST00000229794.4
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr19_+_16186903 | 0.47 |
ENST00000588507.1
|
TPM4
|
tropomyosin 4 |
| chr1_-_182360498 | 0.46 |
ENST00000417584.2
|
GLUL
|
glutamate-ammonia ligase |
| chrX_-_134049233 | 0.46 |
ENST00000370779.4
|
MOSPD1
|
motile sperm domain containing 1 |
| chr1_-_47184723 | 0.46 |
ENST00000371933.3
|
EFCAB14
|
EF-hand calcium binding domain 14 |
| chr20_+_37434329 | 0.45 |
ENST00000299824.1
ENST00000373331.2 |
PPP1R16B
|
protein phosphatase 1, regulatory subunit 16B |
| chr7_-_100808843 | 0.45 |
ENST00000249330.2
|
VGF
|
VGF nerve growth factor inducible |
| chr8_+_97506033 | 0.45 |
ENST00000518385.1
|
SDC2
|
syndecan 2 |
| chr17_+_71161140 | 0.44 |
ENST00000357585.2
|
SSTR2
|
somatostatin receptor 2 |
| chr12_+_57482877 | 0.44 |
ENST00000342556.6
ENST00000357680.4 |
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr19_+_47104553 | 0.43 |
ENST00000598871.1
ENST00000594523.1 |
CALM3
|
calmodulin 3 (phosphorylase kinase, delta) |
| chr1_+_164528866 | 0.42 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr9_+_134165063 | 0.41 |
ENST00000372264.3
|
PPAPDC3
|
phosphatidic acid phosphatase type 2 domain containing 3 |
| chr15_-_75198954 | 0.41 |
ENST00000565772.1
|
FAM219B
|
family with sequence similarity 219, member B |
| chr15_-_48937884 | 0.41 |
ENST00000560355.1
|
FBN1
|
fibrillin 1 |
| chr7_-_11871815 | 0.40 |
ENST00000423059.4
|
THSD7A
|
thrombospondin, type I, domain containing 7A |
| chr1_-_182361327 | 0.39 |
ENST00000331872.6
ENST00000311223.5 |
GLUL
|
glutamate-ammonia ligase |
| chr8_+_142138711 | 0.39 |
ENST00000518347.1
ENST00000262585.2 ENST00000424248.1 ENST00000519811.1 ENST00000520986.1 ENST00000523058.1 |
DENND3
|
DENN/MADD domain containing 3 |
| chr20_+_58179582 | 0.39 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr16_-_30107491 | 0.39 |
ENST00000566134.1
ENST00000565110.1 ENST00000398841.1 ENST00000398838.4 |
YPEL3
|
yippee-like 3 (Drosophila) |
| chr4_+_87856191 | 0.39 |
ENST00000503477.1
|
AFF1
|
AF4/FMR2 family, member 1 |
| chr8_+_142138799 | 0.38 |
ENST00000518668.1
|
DENND3
|
DENN/MADD domain containing 3 |
| chr7_-_105925367 | 0.38 |
ENST00000354289.4
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr15_-_34629922 | 0.37 |
ENST00000559484.1
ENST00000354181.3 ENST00000558589.1 ENST00000458406.2 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr19_-_19051103 | 0.37 |
ENST00000542541.2
ENST00000433218.2 |
HOMER3
|
homer homolog 3 (Drosophila) |
| chr19_+_41107249 | 0.37 |
ENST00000396819.3
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr10_-_103347883 | 0.37 |
ENST00000339310.3
ENST00000370158.3 ENST00000299206.4 ENST00000456836.2 ENST00000413344.1 ENST00000429502.1 ENST00000430045.1 ENST00000370172.1 ENST00000436284.2 ENST00000370162.3 |
POLL
|
polymerase (DNA directed), lambda |
| chr9_+_115513003 | 0.37 |
ENST00000374232.3
|
SNX30
|
sorting nexin family member 30 |
| chr16_-_90085824 | 0.37 |
ENST00000002501.6
|
DBNDD1
|
dysbindin (dystrobrevin binding protein 1) domain containing 1 |
| chr15_-_75199213 | 0.37 |
ENST00000562698.1
|
FAM219B
|
family with sequence similarity 219, member B |
| chr2_+_70485220 | 0.37 |
ENST00000433351.2
ENST00000264441.5 |
PCYOX1
|
prenylcysteine oxidase 1 |
| chr1_-_85930823 | 0.36 |
ENST00000284031.8
ENST00000539042.1 |
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr11_+_71249071 | 0.36 |
ENST00000398534.3
|
KRTAP5-8
|
keratin associated protein 5-8 |
| chr17_-_34122596 | 0.36 |
ENST00000250144.8
|
MMP28
|
matrix metallopeptidase 28 |
| chr2_-_219696519 | 0.36 |
ENST00000545803.1
ENST00000430489.1 ENST00000392098.3 |
PRKAG3
|
protein kinase, AMP-activated, gamma 3 non-catalytic subunit |
| chr5_-_133747589 | 0.35 |
ENST00000458198.2
|
CDKN2AIPNL
|
CDKN2A interacting protein N-terminal like |
| chr1_+_32538492 | 0.35 |
ENST00000336294.5
|
TMEM39B
|
transmembrane protein 39B |
| chr5_-_54281407 | 0.34 |
ENST00000381403.4
|
ESM1
|
endothelial cell-specific molecule 1 |
| chr19_+_45281118 | 0.34 |
ENST00000270279.3
ENST00000341505.4 |
CBLC
|
Cbl proto-oncogene C, E3 ubiquitin protein ligase |
| chr17_-_39324424 | 0.34 |
ENST00000391356.2
|
KRTAP4-3
|
keratin associated protein 4-3 |
| chr17_+_28705921 | 0.34 |
ENST00000225719.4
|
CPD
|
carboxypeptidase D |
| chr17_+_74261413 | 0.34 |
ENST00000587913.1
|
UBALD2
|
UBA-like domain containing 2 |
| chr21_+_38071430 | 0.34 |
ENST00000290399.6
|
SIM2
|
single-minded family bHLH transcription factor 2 |
| chr5_+_176873446 | 0.34 |
ENST00000507881.1
|
PRR7
|
proline rich 7 (synaptic) |
| chr9_+_131218336 | 0.34 |
ENST00000372814.3
|
ODF2
|
outer dense fiber of sperm tails 2 |
| chr4_-_102268484 | 0.33 |
ENST00000394853.4
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr1_+_32538520 | 0.33 |
ENST00000438825.1
ENST00000456834.2 ENST00000373634.4 ENST00000427288.1 |
TMEM39B
|
transmembrane protein 39B |
| chrX_+_118370211 | 0.33 |
ENST00000217971.7
|
PGRMC1
|
progesterone receptor membrane component 1 |
| chr12_+_130646999 | 0.32 |
ENST00000539839.1
ENST00000229030.4 |
FZD10
|
frizzled family receptor 10 |
| chr17_-_27278445 | 0.32 |
ENST00000268756.3
ENST00000584685.1 |
PHF12
|
PHD finger protein 12 |
| chr3_+_5020801 | 0.32 |
ENST00000256495.3
|
BHLHE40
|
basic helix-loop-helix family, member e40 |
| chr13_+_49550015 | 0.32 |
ENST00000492622.2
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr18_+_3448455 | 0.32 |
ENST00000549780.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr4_+_87856129 | 0.32 |
ENST00000395146.4
ENST00000507468.1 |
AFF1
|
AF4/FMR2 family, member 1 |
| chr12_-_59313270 | 0.32 |
ENST00000379141.4
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3 |
| chr14_-_82000140 | 0.32 |
ENST00000555824.1
ENST00000557372.1 ENST00000336735.4 |
SEL1L
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr19_+_16187085 | 0.31 |
ENST00000300933.4
|
TPM4
|
tropomyosin 4 |
| chr20_-_48770174 | 0.30 |
ENST00000341698.2
|
TMEM189-UBE2V1
|
TMEM189-UBE2V1 readthrough |
| chr19_-_41196458 | 0.30 |
ENST00000598779.1
|
NUMBL
|
numb homolog (Drosophila)-like |
| chr2_+_65283506 | 0.30 |
ENST00000377990.2
|
CEP68
|
centrosomal protein 68kDa |
| chr8_+_42995548 | 0.30 |
ENST00000458501.2
ENST00000379644.4 |
HGSNAT
|
heparan-alpha-glucosaminide N-acetyltransferase |
| chr2_-_232791038 | 0.30 |
ENST00000295440.2
ENST00000409852.1 |
NPPC
|
natriuretic peptide C |
| chr4_-_102268628 | 0.30 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr1_+_3388181 | 0.29 |
ENST00000418137.1
ENST00000413250.2 |
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr17_+_39261584 | 0.29 |
ENST00000391415.1
|
KRTAP4-9
|
keratin associated protein 4-9 |
| chr10_-_89623194 | 0.29 |
ENST00000445946.3
|
KLLN
|
killin, p53-regulated DNA replication inhibitor |
| chr10_-_27529486 | 0.29 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr2_+_65283529 | 0.29 |
ENST00000546106.1
ENST00000537589.1 ENST00000260569.4 |
CEP68
|
centrosomal protein 68kDa |
| chr2_+_33359646 | 0.28 |
ENST00000390003.4
ENST00000418533.2 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr3_-_196911002 | 0.28 |
ENST00000452595.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr2_+_33359687 | 0.28 |
ENST00000402934.1
ENST00000404525.1 ENST00000407925.1 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr17_-_39191107 | 0.28 |
ENST00000344363.5
|
KRTAP1-3
|
keratin associated protein 1-3 |
| chr20_-_35274548 | 0.28 |
ENST00000262866.4
|
SLA2
|
Src-like-adaptor 2 |
| chr8_-_59572093 | 0.28 |
ENST00000427130.2
|
NSMAF
|
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr10_+_112257596 | 0.27 |
ENST00000369583.3
|
DUSP5
|
dual specificity phosphatase 5 |
| chr10_+_1102303 | 0.27 |
ENST00000381329.1
|
WDR37
|
WD repeat domain 37 |
| chr1_-_244013384 | 0.27 |
ENST00000366539.1
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr5_+_139027877 | 0.27 |
ENST00000302517.3
|
CXXC5
|
CXXC finger protein 5 |
| chr4_-_80994471 | 0.27 |
ENST00000295465.4
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr7_-_100808394 | 0.27 |
ENST00000445482.2
|
VGF
|
VGF nerve growth factor inducible |
| chr3_-_196910721 | 0.27 |
ENST00000443183.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr10_-_81205373 | 0.27 |
ENST00000372336.3
|
ZCCHC24
|
zinc finger, CCHC domain containing 24 |
| chrX_+_107069063 | 0.27 |
ENST00000262843.6
|
MID2
|
midline 2 |
| chrX_-_40036520 | 0.26 |
ENST00000406200.2
ENST00000378455.4 ENST00000342274.4 |
BCOR
|
BCL6 corepressor |
| chr1_-_91487770 | 0.26 |
ENST00000337393.5
|
ZNF644
|
zinc finger protein 644 |
| chr2_+_16080659 | 0.26 |
ENST00000281043.3
|
MYCN
|
v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
| chr7_-_767249 | 0.26 |
ENST00000403562.1
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr3_-_197024965 | 0.25 |
ENST00000392382.2
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr15_+_74833518 | 0.25 |
ENST00000346246.5
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr17_-_18950310 | 0.25 |
ENST00000573099.1
|
GRAP
|
GRB2-related adaptor protein |
| chr12_-_31744031 | 0.25 |
ENST00000389082.5
|
DENND5B
|
DENN/MADD domain containing 5B |
| chr15_+_25200108 | 0.25 |
ENST00000577949.1
ENST00000338094.6 ENST00000338327.4 ENST00000579070.1 ENST00000577565.1 |
SNURF
SNRPN
|
SNRPN upstream reading frame protein small nuclear ribonucleoprotein polypeptide N |
| chr11_+_62556596 | 0.25 |
ENST00000526546.1
|
TMEM179B
|
transmembrane protein 179B |
| chr14_-_37051798 | 0.24 |
ENST00000258829.5
|
NKX2-8
|
NK2 homeobox 8 |
| chr2_+_232063436 | 0.24 |
ENST00000440107.1
|
ARMC9
|
armadillo repeat containing 9 |
| chr10_+_82298088 | 0.24 |
ENST00000470604.2
|
SH2D4B
|
SH2 domain containing 4B |
| chr19_+_46001697 | 0.24 |
ENST00000451287.2
ENST00000324688.4 |
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr2_+_231577532 | 0.24 |
ENST00000258418.5
|
CAB39
|
calcium binding protein 39 |
| chr11_-_64825993 | 0.24 |
ENST00000340252.4
ENST00000355721.3 ENST00000356632.3 ENST00000355369.2 ENST00000339885.2 ENST00000358658.3 |
NAALADL1
|
N-acetylated alpha-linked acidic dipeptidase-like 1 |
| chr8_+_37654424 | 0.24 |
ENST00000315215.7
|
GPR124
|
G protein-coupled receptor 124 |
| chr1_-_22263790 | 0.24 |
ENST00000374695.3
|
HSPG2
|
heparan sulfate proteoglycan 2 |
| chr3_-_133614297 | 0.24 |
ENST00000486858.1
ENST00000477759.1 |
RAB6B
|
RAB6B, member RAS oncogene family |
| chr2_-_70475701 | 0.24 |
ENST00000282574.4
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chrX_+_9754461 | 0.23 |
ENST00000380913.3
|
SHROOM2
|
shroom family member 2 |
| chr1_+_117910047 | 0.23 |
ENST00000356554.3
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr8_+_86376081 | 0.23 |
ENST00000285379.5
|
CA2
|
carbonic anhydrase II |
| chr22_-_36902522 | 0.23 |
ENST00000397223.4
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr19_-_46418033 | 0.23 |
ENST00000341294.2
|
NANOS2
|
nanos homolog 2 (Drosophila) |
| chr1_-_221915418 | 0.23 |
ENST00000323825.3
ENST00000366899.3 |
DUSP10
|
dual specificity phosphatase 10 |
| chr9_-_79307096 | 0.23 |
ENST00000376717.2
ENST00000223609.6 ENST00000443509.2 |
PRUNE2
|
prune homolog 2 (Drosophila) |
| chr2_+_54683419 | 0.23 |
ENST00000356805.4
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr3_-_196669298 | 0.23 |
ENST00000411704.1
ENST00000452404.2 |
NCBP2
|
nuclear cap binding protein subunit 2, 20kDa |
| chr4_+_95373037 | 0.23 |
ENST00000359265.4
ENST00000437932.1 ENST00000380180.3 ENST00000318007.5 ENST00000450793.1 ENST00000538141.1 ENST00000317968.4 ENST00000512274.1 ENST00000503974.1 ENST00000504489.1 ENST00000542407.1 |
PDLIM5
|
PDZ and LIM domain 5 |
| chr13_+_35516390 | 0.23 |
ENST00000540320.1
ENST00000400445.3 ENST00000310336.4 |
NBEA
|
neurobeachin |
| chr12_+_50017327 | 0.23 |
ENST00000261897.1
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) |
| chr7_-_92855762 | 0.23 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr12_+_50017184 | 0.23 |
ENST00000548825.2
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) |
| chr5_+_76114758 | 0.23 |
ENST00000514165.1
ENST00000296677.4 |
F2RL1
|
coagulation factor II (thrombin) receptor-like 1 |
| chr19_+_1286097 | 0.22 |
ENST00000215368.2
|
EFNA2
|
ephrin-A2 |
| chr6_-_84418841 | 0.22 |
ENST00000369694.2
ENST00000195649.6 |
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr2_-_19558373 | 0.22 |
ENST00000272223.2
|
OSR1
|
odd-skipped related transciption factor 1 |
| chr15_-_52472078 | 0.22 |
ENST00000396335.4
ENST00000560116.1 ENST00000358784.7 |
GNB5
|
guanine nucleotide binding protein (G protein), beta 5 |
| chr6_-_84419101 | 0.22 |
ENST00000520302.1
ENST00000520213.1 ENST00000439399.2 ENST00000428679.2 ENST00000437520.1 |
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr2_+_11052054 | 0.22 |
ENST00000295082.1
|
KCNF1
|
potassium voltage-gated channel, subfamily F, member 1 |
| chr6_-_84418860 | 0.22 |
ENST00000521743.1
|
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr3_+_42727011 | 0.22 |
ENST00000287777.4
|
KLHL40
|
kelch-like family member 40 |
| chr8_-_4851937 | 0.22 |
ENST00000537824.1
ENST00000542608.1 ENST00000539096.1 |
CSMD1
|
CUB and Sushi multiple domains 1 |
| chr11_+_31531291 | 0.22 |
ENST00000350638.5
ENST00000379163.5 ENST00000395934.2 |
ELP4
|
elongator acetyltransferase complex subunit 4 |
| chr2_+_69002052 | 0.21 |
ENST00000497079.1
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr3_-_65583561 | 0.21 |
ENST00000460329.2
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr4_-_83719983 | 0.21 |
ENST00000319540.4
|
SCD5
|
stearoyl-CoA desaturase 5 |
| chr1_+_65210772 | 0.21 |
ENST00000371072.4
ENST00000294428.3 |
RAVER2
|
ribonucleoprotein, PTB-binding 2 |
| chr6_-_30712313 | 0.21 |
ENST00000376377.2
ENST00000259874.5 |
IER3
|
immediate early response 3 |
| chr11_-_17035943 | 0.21 |
ENST00000355661.3
ENST00000532079.1 ENST00000448080.2 ENST00000531066.1 |
PLEKHA7
|
pleckstrin homology domain containing, family A member 7 |
| chr2_-_70475730 | 0.21 |
ENST00000445587.1
ENST00000433529.2 ENST00000415783.2 |
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr15_+_25200074 | 0.21 |
ENST00000390687.4
ENST00000584968.1 ENST00000346403.6 ENST00000554227.2 |
SNRPN
|
small nuclear ribonucleoprotein polypeptide N |
| chrX_+_118370288 | 0.21 |
ENST00000535419.1
|
PGRMC1
|
progesterone receptor membrane component 1 |
| chr11_-_67888881 | 0.21 |
ENST00000356135.5
|
CHKA
|
choline kinase alpha |
| chr8_-_68255912 | 0.21 |
ENST00000262215.3
ENST00000519436.1 |
ARFGEF1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chrX_+_70521584 | 0.20 |
ENST00000373829.3
ENST00000538820.1 |
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2 |
| chr13_-_45150392 | 0.20 |
ENST00000501704.2
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr7_+_128784712 | 0.20 |
ENST00000289407.4
|
TSPAN33
|
tetraspanin 33 |
| chr20_-_33680588 | 0.20 |
ENST00000451813.2
ENST00000432634.2 |
TRPC4AP
|
transient receptor potential cation channel, subfamily C, member 4 associated protein |
| chr16_-_2770216 | 0.20 |
ENST00000302641.3
|
PRSS27
|
protease, serine 27 |
| chrX_-_45710920 | 0.20 |
ENST00000456532.1
|
RP5-1158E12.3
|
RP5-1158E12.3 |
| chr2_-_70475586 | 0.20 |
ENST00000416149.2
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr21_+_34442439 | 0.20 |
ENST00000382348.1
ENST00000333063.5 |
OLIG1
|
oligodendrocyte transcription factor 1 |
| chr13_+_88324870 | 0.20 |
ENST00000325089.6
|
SLITRK5
|
SLIT and NTRK-like family, member 5 |
| chr12_-_113658892 | 0.20 |
ENST00000299732.2
ENST00000416617.2 |
IQCD
|
IQ motif containing D |
| chr10_+_105036909 | 0.20 |
ENST00000369849.4
|
INA
|
internexin neuronal intermediate filament protein, alpha |
| chr19_+_38755203 | 0.20 |
ENST00000587090.1
ENST00000454580.3 |
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NHLH1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.3 | 1.7 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.2 | 0.9 | GO:0061227 | intermediate mesoderm development(GO:0048389) pattern specification involved in mesonephros development(GO:0061227) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.2 | 0.6 | GO:0021571 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.2 | 0.6 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.2 | 1.0 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.2 | 0.9 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.2 | 0.5 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 0.5 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 1.9 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 0.5 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 | 1.0 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.1 | 1.0 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.1 | 0.3 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 0.3 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.1 | 3.4 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 1.0 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.7 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.4 | GO:0046833 | snRNA export from nucleus(GO:0006408) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.1 | 0.4 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.1 | 0.3 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.2 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 0.3 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.4 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 0.4 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 0.2 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 0.4 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 0.3 | GO:1900194 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.4 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 0.2 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.2 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.1 | 0.5 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.1 | 0.4 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.1 | 0.3 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.4 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.3 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.3 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.1 | 0.4 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 0.5 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 | 1.2 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.2 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 0.6 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.6 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.2 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.7 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.3 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.0 | 0.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.2 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.0 | 0.2 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.1 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.0 | 0.9 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 1.1 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.5 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.0 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:1904397 | negative regulation of neuromuscular junction development(GO:1904397) |
| 0.0 | 0.2 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.2 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.5 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.5 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.3 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.1 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.1 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.0 | 0.4 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.0 | 0.1 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.3 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.7 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.3 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.3 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.1 | GO:0006311 | meiotic gene conversion(GO:0006311) |
| 0.0 | 0.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.6 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.1 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.1 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.4 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.1 | GO:0060380 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.0 | 0.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 1.7 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.1 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.6 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.3 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.1 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.2 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.4 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.0 | 0.8 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.2 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 0.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.1 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.2 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.4 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.4 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.4 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.8 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.0 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.0 | 0.1 | GO:1904263 | cellular response to leucine(GO:0071233) positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.1 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 0.0 | 0.1 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 0.0 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0005940 | septin ring(GO:0005940) septin collar(GO:0032173) |
| 0.1 | 0.5 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.1 | 0.5 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.1 | 0.4 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.4 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.1 | 0.6 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 1.0 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 0.3 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 1.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.4 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.8 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.7 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 1.0 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.3 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.9 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.4 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 1.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 3.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 1.8 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.4 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 0.7 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 1.7 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 0.5 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.1 | 0.5 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 0.6 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 3.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.4 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 1.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.3 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 0.2 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.1 | 0.4 | GO:0044594 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.1 | 0.6 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.2 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.2 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 1.0 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.5 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 0.4 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.7 | GO:0039706 | co-receptor binding(GO:0039706) BMP receptor binding(GO:0070700) |
| 0.0 | 0.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.5 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.4 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.2 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.6 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 1.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.2 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.3 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.4 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.4 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.4 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.1 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.0 | 0.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 1.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.4 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 1.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 0.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.1 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.5 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.0 | 0.3 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.5 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 1.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.6 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.7 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.1 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.6 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 1.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 3.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 1.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |