Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
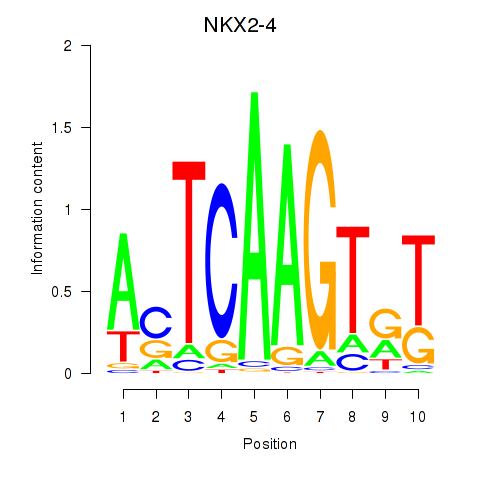
Results for NKX2-4
Z-value: 0.57
Transcription factors associated with NKX2-4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-4
|
ENSG00000125816.4 | NK2 homeobox 4 |
Activity profile of NKX2-4 motif
Sorted Z-values of NKX2-4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_102651343 | 1.73 |
ENST00000279441.4
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 (stromelysin 2) |
| chr15_+_67430339 | 1.26 |
ENST00000439724.3
|
SMAD3
|
SMAD family member 3 |
| chr7_-_47579188 | 1.25 |
ENST00000398879.1
ENST00000355730.3 ENST00000442536.2 ENST00000458317.2 |
TNS3
|
tensin 3 |
| chr12_-_71182695 | 1.22 |
ENST00000342084.4
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr3_-_107777208 | 1.22 |
ENST00000398258.3
|
CD47
|
CD47 molecule |
| chr5_-_61031495 | 1.15 |
ENST00000506550.1
ENST00000512882.2 |
CTD-2170G1.2
|
CTD-2170G1.2 |
| chr2_+_161993465 | 1.09 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr1_-_85870177 | 0.96 |
ENST00000542148.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr7_+_134551583 | 0.95 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chrX_-_154563889 | 0.91 |
ENST00000369449.2
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr7_-_115608304 | 0.83 |
ENST00000457268.1
|
TFEC
|
transcription factor EC |
| chr7_+_129932974 | 0.82 |
ENST00000445470.2
ENST00000222482.4 ENST00000492072.1 ENST00000473956.1 ENST00000493259.1 ENST00000486598.1 |
CPA4
|
carboxypeptidase A4 |
| chr19_+_45504688 | 0.81 |
ENST00000221452.8
ENST00000540120.1 ENST00000505236.1 |
RELB
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr4_-_80994619 | 0.80 |
ENST00000404191.1
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr13_-_43566301 | 0.79 |
ENST00000398762.3
ENST00000313640.7 ENST00000313624.7 |
EPSTI1
|
epithelial stromal interaction 1 (breast) |
| chr1_+_144989309 | 0.79 |
ENST00000596396.1
|
AL590452.1
|
Uncharacterized protein |
| chr1_+_205225319 | 0.75 |
ENST00000329800.7
|
TMCC2
|
transmembrane and coiled-coil domain family 2 |
| chr1_-_89591749 | 0.72 |
ENST00000370466.3
|
GBP2
|
guanylate binding protein 2, interferon-inducible |
| chr12_+_10658201 | 0.70 |
ENST00000322446.3
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr2_+_161993412 | 0.70 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr18_-_33702078 | 0.67 |
ENST00000586829.1
|
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr4_-_80994471 | 0.66 |
ENST00000295465.4
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr19_+_10765699 | 0.65 |
ENST00000590009.1
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chrX_+_144908928 | 0.64 |
ENST00000408967.2
|
TMEM257
|
transmembrane protein 257 |
| chr22_+_41697520 | 0.64 |
ENST00000352645.4
|
ZC3H7B
|
zinc finger CCCH-type containing 7B |
| chr16_+_1730338 | 0.62 |
ENST00000566691.1
ENST00000382710.4 |
HN1L
|
hematological and neurological expressed 1-like |
| chr21_+_40177143 | 0.61 |
ENST00000360214.3
|
ETS2
|
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
| chr4_+_86748898 | 0.58 |
ENST00000509300.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr6_+_83072923 | 0.56 |
ENST00000535040.1
|
TPBG
|
trophoblast glycoprotein |
| chr15_-_55541227 | 0.56 |
ENST00000566877.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr2_-_37899323 | 0.55 |
ENST00000295324.3
ENST00000457889.1 |
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr12_-_48500085 | 0.54 |
ENST00000549518.1
|
SENP1
|
SUMO1/sentrin specific peptidase 1 |
| chr17_-_40264692 | 0.53 |
ENST00000591220.1
ENST00000251642.3 |
DHX58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr4_+_96012614 | 0.52 |
ENST00000264568.4
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr2_-_202562716 | 0.52 |
ENST00000428900.2
|
MPP4
|
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr11_-_105892937 | 0.51 |
ENST00000301919.4
ENST00000534458.1 ENST00000530108.1 ENST00000530788.1 |
MSANTD4
|
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
| chr14_+_51955831 | 0.49 |
ENST00000356218.4
|
FRMD6
|
FERM domain containing 6 |
| chr4_+_86749045 | 0.49 |
ENST00000514229.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr19_-_55669093 | 0.49 |
ENST00000344887.5
|
TNNI3
|
troponin I type 3 (cardiac) |
| chr7_-_92463210 | 0.48 |
ENST00000265734.4
|
CDK6
|
cyclin-dependent kinase 6 |
| chr8_+_104310661 | 0.47 |
ENST00000522566.1
|
FZD6
|
frizzled family receptor 6 |
| chr7_+_151038850 | 0.47 |
ENST00000355851.4
ENST00000566856.1 ENST00000470229.1 |
NUB1
|
negative regulator of ubiquitin-like proteins 1 |
| chr5_-_13944652 | 0.46 |
ENST00000265104.4
|
DNAH5
|
dynein, axonemal, heavy chain 5 |
| chr4_-_103746683 | 0.44 |
ENST00000504211.1
ENST00000508476.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr6_+_37225540 | 0.43 |
ENST00000373491.3
|
TBC1D22B
|
TBC1 domain family, member 22B |
| chr6_+_126240442 | 0.42 |
ENST00000448104.1
ENST00000438495.1 ENST00000444128.1 |
NCOA7
|
nuclear receptor coactivator 7 |
| chr3_+_173116225 | 0.41 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr6_+_83073334 | 0.41 |
ENST00000369750.3
|
TPBG
|
trophoblast glycoprotein |
| chr11_-_85780853 | 0.40 |
ENST00000531930.1
ENST00000528398.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr2_+_162016916 | 0.40 |
ENST00000405852.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr11_+_35965531 | 0.40 |
ENST00000528989.1
ENST00000524419.1 ENST00000315571.5 |
LDLRAD3
|
low density lipoprotein receptor class A domain containing 3 |
| chr6_+_153552455 | 0.39 |
ENST00000392385.2
|
AL590867.1
|
Uncharacterized protein; cDNA FLJ59044, highly similar to LINE-1 reverse transcriptase homolog |
| chr19_+_36235964 | 0.38 |
ENST00000587708.2
|
PSENEN
|
presenilin enhancer gamma secretase subunit |
| chr11_-_85779971 | 0.37 |
ENST00000393346.3
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr2_-_197226875 | 0.37 |
ENST00000409111.1
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr2_-_175462456 | 0.37 |
ENST00000409891.1
ENST00000410117.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr3_+_30647994 | 0.37 |
ENST00000295754.5
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chr8_+_31496809 | 0.36 |
ENST00000518104.1
ENST00000519301.1 |
NRG1
|
neuregulin 1 |
| chr12_+_75874460 | 0.35 |
ENST00000266659.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr5_-_59481406 | 0.35 |
ENST00000546160.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr3_+_173302222 | 0.35 |
ENST00000361589.4
|
NLGN1
|
neuroligin 1 |
| chr18_+_55816546 | 0.35 |
ENST00000435432.2
ENST00000357895.5 ENST00000586263.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr1_-_156721502 | 0.35 |
ENST00000357325.5
|
HDGF
|
hepatoma-derived growth factor |
| chr2_-_100759037 | 0.34 |
ENST00000317233.4
ENST00000423966.1 ENST00000416492.1 |
AFF3
|
AF4/FMR2 family, member 3 |
| chr5_-_160279207 | 0.34 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr4_-_103747011 | 0.34 |
ENST00000350435.7
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr17_+_42923686 | 0.34 |
ENST00000591513.1
|
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B |
| chr20_-_21494654 | 0.34 |
ENST00000377142.4
|
NKX2-2
|
NK2 homeobox 2 |
| chr4_-_103746924 | 0.34 |
ENST00000505207.1
ENST00000502404.1 ENST00000507845.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr10_+_60094735 | 0.33 |
ENST00000373910.4
|
UBE2D1
|
ubiquitin-conjugating enzyme E2D 1 |
| chr19_+_7733929 | 0.33 |
ENST00000221515.2
|
RETN
|
resistin |
| chr5_-_98262240 | 0.32 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr1_-_217250231 | 0.31 |
ENST00000493748.1
ENST00000463665.1 |
ESRRG
|
estrogen-related receptor gamma |
| chr1_-_13390765 | 0.31 |
ENST00000357367.2
|
PRAMEF8
|
PRAME family member 8 |
| chr6_+_45390222 | 0.31 |
ENST00000359524.5
|
RUNX2
|
runt-related transcription factor 2 |
| chr5_+_137419581 | 0.30 |
ENST00000506684.1
ENST00000504809.1 ENST00000398754.1 |
WNT8A
|
wingless-type MMTV integration site family, member 8A |
| chr11_-_85780086 | 0.29 |
ENST00000532317.1
ENST00000528256.1 ENST00000526033.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr15_-_88799661 | 0.29 |
ENST00000360948.2
ENST00000357724.2 ENST00000355254.2 ENST00000317501.3 |
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr12_+_124069070 | 0.28 |
ENST00000262225.3
ENST00000438031.2 |
TMED2
|
transmembrane emp24 domain trafficking protein 2 |
| chr6_+_12007897 | 0.28 |
ENST00000437559.1
|
RP11-456H18.2
|
RP11-456H18.2 |
| chr5_+_154135029 | 0.28 |
ENST00000518297.1
|
LARP1
|
La ribonucleoprotein domain family, member 1 |
| chr1_+_175036966 | 0.28 |
ENST00000239462.4
|
TNN
|
tenascin N |
| chr10_+_105005644 | 0.28 |
ENST00000441178.2
|
RP11-332O19.5
|
ribulose-5-phosphate-3-epimerase-like 1 |
| chr4_+_71063641 | 0.28 |
ENST00000514097.1
|
ODAM
|
odontogenic, ameloblast asssociated |
| chr3_-_88108212 | 0.27 |
ENST00000482016.1
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr1_-_89458287 | 0.27 |
ENST00000370485.2
|
CCBL2
|
cysteine conjugate-beta lyase 2 |
| chr1_-_85040090 | 0.27 |
ENST00000370630.5
|
CTBS
|
chitobiase, di-N-acetyl- |
| chr4_+_71263599 | 0.26 |
ENST00000399575.2
|
PROL1
|
proline rich, lacrimal 1 |
| chr1_-_47407097 | 0.26 |
ENST00000457840.2
|
CYP4A11
|
cytochrome P450, family 4, subfamily A, polypeptide 11 |
| chr21_-_15918618 | 0.26 |
ENST00000400564.1
ENST00000400566.1 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chrX_-_30993201 | 0.25 |
ENST00000288422.2
ENST00000378932.2 |
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr2_+_183982238 | 0.25 |
ENST00000442895.2
ENST00000446612.1 ENST00000409798.1 |
NUP35
|
nucleoporin 35kDa |
| chr20_-_43280325 | 0.25 |
ENST00000537820.1
|
ADA
|
adenosine deaminase |
| chr11_+_60699222 | 0.25 |
ENST00000536409.1
|
TMEM132A
|
transmembrane protein 132A |
| chrX_-_138914394 | 0.25 |
ENST00000327569.3
ENST00000361648.2 ENST00000370543.1 ENST00000359686.2 |
ATP11C
|
ATPase, class VI, type 11C |
| chr16_-_21877629 | 0.24 |
ENST00000544693.1
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr1_-_204183071 | 0.24 |
ENST00000308302.3
|
GOLT1A
|
golgi transport 1A |
| chr10_-_48055018 | 0.24 |
ENST00000426610.2
|
ASAH2C
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2C |
| chr18_+_11851383 | 0.23 |
ENST00000526991.2
|
CHMP1B
|
charged multivesicular body protein 1B |
| chr7_+_116139424 | 0.23 |
ENST00000222693.4
|
CAV2
|
caveolin 2 |
| chrX_+_70443050 | 0.23 |
ENST00000361726.6
|
GJB1
|
gap junction protein, beta 1, 32kDa |
| chr21_-_35884573 | 0.23 |
ENST00000399286.2
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1 |
| chr22_-_50524298 | 0.23 |
ENST00000311597.5
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr1_-_95391315 | 0.23 |
ENST00000545882.1
ENST00000415017.1 |
CNN3
|
calponin 3, acidic |
| chr11_+_60524426 | 0.23 |
ENST00000528170.1
ENST00000337911.4 ENST00000405633.3 |
MS4A15
|
membrane-spanning 4-domains, subfamily A, member 15 |
| chr10_+_98592674 | 0.22 |
ENST00000356016.3
ENST00000371097.4 |
LCOR
|
ligand dependent nuclear receptor corepressor |
| chr15_+_63354769 | 0.22 |
ENST00000558910.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr1_-_241520525 | 0.22 |
ENST00000366565.1
|
RGS7
|
regulator of G-protein signaling 7 |
| chr10_-_69455873 | 0.22 |
ENST00000433211.2
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3 |
| chr6_-_152639479 | 0.22 |
ENST00000356820.4
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr2_+_37571845 | 0.22 |
ENST00000537448.1
|
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr16_+_81272287 | 0.21 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr2_+_162272605 | 0.21 |
ENST00000389554.3
|
TBR1
|
T-box, brain, 1 |
| chr8_+_66955648 | 0.21 |
ENST00000522619.1
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr19_-_20748541 | 0.21 |
ENST00000427401.4
ENST00000594419.1 |
ZNF737
|
zinc finger protein 737 |
| chr1_-_205419053 | 0.20 |
ENST00000367154.1
|
LEMD1
|
LEM domain containing 1 |
| chr17_+_60501228 | 0.20 |
ENST00000311506.5
|
METTL2A
|
methyltransferase like 2A |
| chr3_+_15045419 | 0.20 |
ENST00000406272.2
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr15_+_63334831 | 0.20 |
ENST00000288398.6
ENST00000358278.3 ENST00000560445.1 ENST00000403994.3 ENST00000357980.4 ENST00000559556.1 ENST00000559397.1 ENST00000267996.7 ENST00000560970.1 |
TPM1
|
tropomyosin 1 (alpha) |
| chr11_+_61717842 | 0.20 |
ENST00000449131.2
|
BEST1
|
bestrophin 1 |
| chr10_+_99332529 | 0.20 |
ENST00000455090.1
|
ANKRD2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr11_-_119993979 | 0.20 |
ENST00000524816.3
ENST00000525327.1 |
TRIM29
|
tripartite motif containing 29 |
| chr6_-_152958521 | 0.19 |
ENST00000367255.5
ENST00000265368.4 ENST00000448038.1 ENST00000341594.5 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr10_-_61495760 | 0.19 |
ENST00000395347.1
|
SLC16A9
|
solute carrier family 16, member 9 |
| chr8_+_11141925 | 0.19 |
ENST00000221086.3
|
MTMR9
|
myotubularin related protein 9 |
| chr10_+_21823243 | 0.19 |
ENST00000307729.7
ENST00000377091.2 |
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr1_-_21606013 | 0.18 |
ENST00000357071.4
|
ECE1
|
endothelin converting enzyme 1 |
| chr16_+_22516172 | 0.18 |
ENST00000543407.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr3_+_137906154 | 0.18 |
ENST00000466749.1
ENST00000358441.2 ENST00000489213.1 |
ARMC8
|
armadillo repeat containing 8 |
| chr13_-_108870623 | 0.18 |
ENST00000405925.1
|
LIG4
|
ligase IV, DNA, ATP-dependent |
| chr14_-_71067360 | 0.18 |
ENST00000554963.1
ENST00000430055.2 ENST00000440435.2 ENST00000256379.5 |
MED6
|
mediator complex subunit 6 |
| chr8_+_37594103 | 0.18 |
ENST00000397228.2
|
ERLIN2
|
ER lipid raft associated 2 |
| chr16_+_77822427 | 0.18 |
ENST00000302536.2
|
VAT1L
|
vesicle amine transport 1-like |
| chr6_-_131277510 | 0.18 |
ENST00000525193.1
ENST00000527659.1 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr12_+_21168630 | 0.18 |
ENST00000421593.2
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr10_-_121296045 | 0.18 |
ENST00000392865.1
|
RGS10
|
regulator of G-protein signaling 10 |
| chr4_-_87028478 | 0.18 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr2_-_209054709 | 0.17 |
ENST00000449053.1
ENST00000451346.1 ENST00000341287.4 |
C2orf80
|
chromosome 2 open reading frame 80 |
| chr5_+_147443534 | 0.17 |
ENST00000398454.1
ENST00000359874.3 ENST00000508733.1 ENST00000256084.7 |
SPINK5
|
serine peptidase inhibitor, Kazal type 5 |
| chr7_+_143174966 | 0.17 |
ENST00000408916.1
|
TAS2R41
|
taste receptor, type 2, member 41 |
| chr1_-_241520385 | 0.17 |
ENST00000366564.1
|
RGS7
|
regulator of G-protein signaling 7 |
| chr4_+_111397216 | 0.17 |
ENST00000265162.5
|
ENPEP
|
glutamyl aminopeptidase (aminopeptidase A) |
| chr10_+_81272287 | 0.17 |
ENST00000520547.2
|
EIF5AL1
|
eukaryotic translation initiation factor 5A-like 1 |
| chrX_+_78200913 | 0.17 |
ENST00000171757.2
|
P2RY10
|
purinergic receptor P2Y, G-protein coupled, 10 |
| chr6_-_134861089 | 0.17 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr4_-_174320687 | 0.17 |
ENST00000296506.3
|
SCRG1
|
stimulator of chondrogenesis 1 |
| chr1_-_100643765 | 0.17 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr4_+_39699664 | 0.17 |
ENST00000261427.5
ENST00000510934.1 ENST00000295963.6 |
UBE2K
|
ubiquitin-conjugating enzyme E2K |
| chr14_-_104028595 | 0.17 |
ENST00000337322.4
ENST00000445922.2 |
BAG5
|
BCL2-associated athanogene 5 |
| chr5_-_34919094 | 0.17 |
ENST00000341754.4
|
RAD1
|
RAD1 homolog (S. pombe) |
| chr10_+_52094298 | 0.16 |
ENST00000595931.1
|
AC069547.1
|
HCG1745369; PRO3073; Uncharacterized protein |
| chr1_-_42801540 | 0.16 |
ENST00000372573.1
|
FOXJ3
|
forkhead box J3 |
| chr19_-_49016418 | 0.16 |
ENST00000270238.3
|
LMTK3
|
lemur tyrosine kinase 3 |
| chrX_+_78200829 | 0.16 |
ENST00000544091.1
|
P2RY10
|
purinergic receptor P2Y, G-protein coupled, 10 |
| chr2_-_37544209 | 0.16 |
ENST00000234179.2
|
PRKD3
|
protein kinase D3 |
| chr3_+_189349162 | 0.16 |
ENST00000264731.3
ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63
|
tumor protein p63 |
| chr13_-_108867846 | 0.16 |
ENST00000442234.1
|
LIG4
|
ligase IV, DNA, ATP-dependent |
| chr2_-_49381646 | 0.16 |
ENST00000346173.3
ENST00000406846.2 |
FSHR
|
follicle stimulating hormone receptor |
| chr16_-_3450963 | 0.15 |
ENST00000573327.1
ENST00000571906.1 ENST00000573830.1 ENST00000439568.2 ENST00000422427.2 ENST00000304926.3 ENST00000396852.4 |
ZSCAN32
|
zinc finger and SCAN domain containing 32 |
| chr12_-_322504 | 0.15 |
ENST00000424061.2
|
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr6_+_12958137 | 0.15 |
ENST00000457702.2
ENST00000379345.2 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr11_-_10715502 | 0.15 |
ENST00000547195.1
|
MRVI1
|
murine retrovirus integration site 1 homolog |
| chr1_-_155271213 | 0.15 |
ENST00000342741.4
|
PKLR
|
pyruvate kinase, liver and RBC |
| chr8_+_26247878 | 0.15 |
ENST00000518611.1
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr14_-_81893734 | 0.15 |
ENST00000555447.1
|
STON2
|
stonin 2 |
| chr6_+_12007963 | 0.15 |
ENST00000607445.1
|
RP11-456H18.2
|
RP11-456H18.2 |
| chr4_-_175443943 | 0.15 |
ENST00000296522.6
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr18_+_21032781 | 0.15 |
ENST00000339486.3
|
RIOK3
|
RIO kinase 3 |
| chr8_-_6914251 | 0.15 |
ENST00000330590.2
|
DEFA5
|
defensin, alpha 5, Paneth cell-specific |
| chr2_+_204571198 | 0.15 |
ENST00000374481.3
ENST00000458610.2 ENST00000324106.8 |
CD28
|
CD28 molecule |
| chr18_+_9136758 | 0.15 |
ENST00000383440.2
ENST00000262126.4 ENST00000577992.1 |
ANKRD12
|
ankyrin repeat domain 12 |
| chr3_+_4535155 | 0.14 |
ENST00000544951.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr12_+_8975061 | 0.14 |
ENST00000299698.7
|
A2ML1
|
alpha-2-macroglobulin-like 1 |
| chr6_-_39282329 | 0.14 |
ENST00000373231.4
|
KCNK17
|
potassium channel, subfamily K, member 17 |
| chrX_+_49020882 | 0.14 |
ENST00000454342.1
|
MAGIX
|
MAGI family member, X-linked |
| chr15_-_86338100 | 0.14 |
ENST00000536947.1
|
KLHL25
|
kelch-like family member 25 |
| chr10_-_104001231 | 0.14 |
ENST00000370002.3
|
PITX3
|
paired-like homeodomain 3 |
| chr8_+_41347915 | 0.14 |
ENST00000518270.1
ENST00000520817.1 |
GOLGA7
|
golgin A7 |
| chr11_-_19223523 | 0.14 |
ENST00000265968.3
|
CSRP3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr11_+_14926543 | 0.14 |
ENST00000523376.1
|
CALCB
|
calcitonin-related polypeptide beta |
| chrX_-_110655391 | 0.14 |
ENST00000356915.2
ENST00000356220.3 |
DCX
|
doublecortin |
| chr19_-_10047219 | 0.14 |
ENST00000264833.4
|
OLFM2
|
olfactomedin 2 |
| chr13_-_96705624 | 0.14 |
ENST00000376747.3
ENST00000376712.4 ENST00000397618.3 ENST00000376714.3 |
UGGT2
|
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr5_+_161494770 | 0.14 |
ENST00000414552.2
ENST00000361925.4 |
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr3_+_68053359 | 0.14 |
ENST00000478136.1
|
FAM19A1
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A1 |
| chr1_+_152974218 | 0.14 |
ENST00000331860.3
ENST00000443178.1 ENST00000295367.4 |
SPRR3
|
small proline-rich protein 3 |
| chr6_+_159071015 | 0.13 |
ENST00000360448.3
|
SYTL3
|
synaptotagmin-like 3 |
| chr11_+_77774897 | 0.13 |
ENST00000281030.2
|
THRSP
|
thyroid hormone responsive |
| chr14_+_22356029 | 0.13 |
ENST00000390437.2
|
TRAV12-2
|
T cell receptor alpha variable 12-2 |
| chr17_+_28804380 | 0.13 |
ENST00000225724.5
ENST00000451249.2 ENST00000467337.2 ENST00000581721.1 ENST00000414833.2 |
GOSR1
|
golgi SNAP receptor complex member 1 |
| chr11_-_10715287 | 0.13 |
ENST00000423302.2
|
MRVI1
|
murine retrovirus integration site 1 homolog |
| chr14_-_96830207 | 0.13 |
ENST00000359933.4
|
ATG2B
|
autophagy related 2B |
| chr2_+_219110149 | 0.13 |
ENST00000456575.1
|
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr13_+_111767650 | 0.13 |
ENST00000449979.1
ENST00000370623.3 |
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr1_+_15986364 | 0.13 |
ENST00000345034.1
|
RSC1A1
|
regulatory solute carrier protein, family 1, member 1 |
| chr19_+_52264449 | 0.13 |
ENST00000599326.1
ENST00000598953.1 |
FPR2
|
formyl peptide receptor 2 |
| chr14_-_23058063 | 0.13 |
ENST00000538631.1
ENST00000543337.1 ENST00000250498.4 |
DAD1
|
defender against cell death 1 |
| chr6_-_9933500 | 0.13 |
ENST00000492169.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr18_-_28622699 | 0.13 |
ENST00000360428.4
|
DSC3
|
desmocollin 3 |
| chr6_-_39282221 | 0.13 |
ENST00000453413.2
|
KCNK17
|
potassium channel, subfamily K, member 17 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.3 | 1.3 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.3 | 0.8 | GO:0099545 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.2 | 1.2 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.2 | 0.7 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.2 | 1.1 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 0.4 | GO:0002661 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.1 | 0.5 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.1 | 0.3 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 0.3 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.1 | 0.3 | GO:2000870 | positive regulation of female gonad development(GO:2000196) regulation of progesterone secretion(GO:2000870) |
| 0.1 | 0.5 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.6 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 0.3 | GO:0014034 | neural crest cell fate commitment(GO:0014034) |
| 0.1 | 0.3 | GO:0048687 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.1 | 0.6 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.4 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.1 | 0.3 | GO:0046103 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.1 | 0.2 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.1 | 0.3 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.5 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.3 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.1 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 1.0 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.1 | 1.2 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 0.5 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.9 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.1 | 0.3 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 | 0.2 | GO:0051598 | meiotic recombination checkpoint(GO:0051598) |
| 0.1 | 0.5 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.1 | 0.4 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 0.4 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.8 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.1 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.0 | 0.3 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.2 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.1 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.0 | 0.1 | GO:0002325 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.0 | 0.2 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.2 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.0 | 0.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.4 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.7 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 0.0 | 0.1 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.0 | 0.6 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.2 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.8 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.2 | GO:0044789 | receptor-mediated endocytosis of virus by host cell(GO:0019065) modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.2 | GO:0042640 | anagen(GO:0042640) |
| 0.0 | 0.1 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.1 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.3 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.2 | GO:0061083 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.2 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.0 | 0.4 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.2 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.2 | GO:0002005 | angiotensin catabolic process in blood(GO:0002005) |
| 0.0 | 0.2 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.5 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0051941 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.0 | 0.2 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 1.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 1.2 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.2 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.1 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.0 | 0.2 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.1 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.1 | GO:0032811 | negative regulation of epinephrine secretion(GO:0032811) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.0 | 1.1 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.2 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.1 | GO:1902748 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.2 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.4 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 1.6 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.1 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.0 | 0.2 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.1 | GO:2000314 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.0 | 0.2 | GO:0002863 | positive regulation of inflammatory response to antigenic stimulus(GO:0002863) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.2 | GO:0072584 | caveolin-mediated endocytosis(GO:0072584) |
| 0.0 | 0.3 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.7 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.0 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.3 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.0 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.2 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.2 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.0 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.2 | 1.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.5 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.8 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 1.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.4 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.7 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.4 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.5 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.4 | GO:0032059 | muscle thin filament tropomyosin(GO:0005862) bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 1.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.0 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 1.3 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.1 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.3 | 1.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.2 | 0.5 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 1.0 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 1.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.4 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.1 | 0.4 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 0.5 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 1.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.2 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 0.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.5 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.5 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.3 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 1.2 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.0 | 1.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.5 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.3 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.0 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 1.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.2 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.3 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.0 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 2.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.3 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 1.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.0 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 2.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 1.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 1.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.4 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.4 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 1.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 1.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.7 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.0 | 0.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |