Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
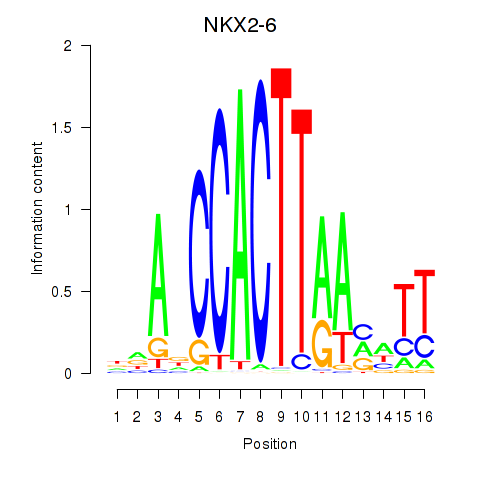
Results for NKX2-6
Z-value: 0.91
Transcription factors associated with NKX2-6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-6
|
ENSG00000180053.6 | NK2 homeobox 6 |
Activity profile of NKX2-6 motif
Sorted Z-values of NKX2-6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_113354341 | 4.65 |
ENST00000553152.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr10_-_6019984 | 4.07 |
ENST00000525219.2
|
IL15RA
|
interleukin 15 receptor, alpha |
| chr10_-_6019552 | 3.94 |
ENST00000379977.3
ENST00000397251.3 ENST00000397248.2 |
IL15RA
|
interleukin 15 receptor, alpha |
| chr4_+_89299885 | 3.83 |
ENST00000380265.5
ENST00000273960.3 |
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr3_-_123339343 | 3.25 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr3_-_123339418 | 3.22 |
ENST00000583087.1
|
MYLK
|
myosin light chain kinase |
| chr3_-_127541194 | 2.76 |
ENST00000453507.2
|
MGLL
|
monoglyceride lipase |
| chr10_-_6019455 | 2.62 |
ENST00000530685.1
ENST00000397255.3 ENST00000379971.1 ENST00000528354.1 ENST00000397250.2 ENST00000429135.2 |
IL15RA
|
interleukin 15 receptor, alpha |
| chr12_+_56075330 | 2.34 |
ENST00000394252.3
|
METTL7B
|
methyltransferase like 7B |
| chr1_+_205225319 | 2.15 |
ENST00000329800.7
|
TMCC2
|
transmembrane and coiled-coil domain family 2 |
| chr2_-_100721923 | 1.89 |
ENST00000356421.2
|
AFF3
|
AF4/FMR2 family, member 3 |
| chr4_+_70916119 | 1.84 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chr2_+_102928009 | 1.81 |
ENST00000404917.2
ENST00000447231.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr9_+_96846740 | 1.50 |
ENST00000288976.3
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1 |
| chr12_-_89919765 | 1.46 |
ENST00000541909.1
ENST00000313546.3 |
POC1B
|
POC1 centriolar protein B |
| chr17_-_55911970 | 1.39 |
ENST00000581805.1
ENST00000580960.1 |
RP11-60A24.3
|
RP11-60A24.3 |
| chr2_-_100721178 | 1.37 |
ENST00000409236.2
|
AFF3
|
AF4/FMR2 family, member 3 |
| chr12_-_56236690 | 1.37 |
ENST00000322569.4
|
MMP19
|
matrix metallopeptidase 19 |
| chr8_+_27629459 | 1.31 |
ENST00000523566.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr3_-_121379739 | 1.29 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr19_-_44174330 | 1.24 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr1_+_1243947 | 1.23 |
ENST00000379031.5
|
PUSL1
|
pseudouridylate synthase-like 1 |
| chr1_+_144989309 | 1.23 |
ENST00000596396.1
|
AL590452.1
|
Uncharacterized protein |
| chr5_-_59481406 | 1.21 |
ENST00000546160.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr12_-_89919965 | 1.19 |
ENST00000548729.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr15_-_55563072 | 1.17 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr19_-_44174305 | 1.10 |
ENST00000601723.1
ENST00000339082.3 |
PLAUR
|
plasminogen activator, urokinase receptor |
| chr7_-_107880508 | 1.02 |
ENST00000425651.2
|
NRCAM
|
neuronal cell adhesion molecule |
| chr7_+_75544466 | 0.99 |
ENST00000421059.1
ENST00000394893.1 ENST00000412521.1 ENST00000414186.1 |
POR
|
P450 (cytochrome) oxidoreductase |
| chr11_+_94277017 | 0.99 |
ENST00000358752.2
|
FUT4
|
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) |
| chr12_-_89920030 | 0.95 |
ENST00000413530.1
ENST00000547474.1 |
GALNT4
POC1B-GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) POC1B-GALNT4 readthrough |
| chr5_+_52083730 | 0.95 |
ENST00000282588.6
ENST00000274311.2 |
ITGA1
PELO
|
integrin, alpha 1 pelota homolog (Drosophila) |
| chrX_+_30261847 | 0.91 |
ENST00000378981.3
ENST00000397550.1 |
MAGEB1
|
melanoma antigen family B, 1 |
| chr4_-_79860506 | 0.91 |
ENST00000295462.3
ENST00000380645.4 ENST00000512733.1 |
PAQR3
|
progestin and adipoQ receptor family member III |
| chr2_+_70142189 | 0.89 |
ENST00000264444.2
|
MXD1
|
MAX dimerization protein 1 |
| chr3_+_171561127 | 0.84 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr3_+_100428268 | 0.78 |
ENST00000240851.4
|
TFG
|
TRK-fused gene |
| chr1_+_156338993 | 0.78 |
ENST00000368249.1
ENST00000368246.2 ENST00000537040.1 ENST00000400992.2 ENST00000255013.3 ENST00000451864.2 |
RHBG
|
Rh family, B glycoprotein (gene/pseudogene) |
| chr2_-_69180012 | 0.77 |
ENST00000481498.1
|
GKN2
|
gastrokine 2 |
| chr3_+_100428316 | 0.76 |
ENST00000479672.1
ENST00000476228.1 ENST00000463568.1 |
TFG
|
TRK-fused gene |
| chr14_+_52164820 | 0.76 |
ENST00000554167.1
|
FRMD6
|
FERM domain containing 6 |
| chr16_+_78056412 | 0.75 |
ENST00000299642.4
ENST00000575655.1 |
CLEC3A
|
C-type lectin domain family 3, member A |
| chr17_-_29624343 | 0.74 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr10_+_121652204 | 0.73 |
ENST00000369075.3
ENST00000543134.1 |
SEC23IP
|
SEC23 interacting protein |
| chr13_+_44453969 | 0.73 |
ENST00000325686.6
|
LACC1
|
laccase (multicopper oxidoreductase) domain containing 1 |
| chr12_-_71182695 | 0.69 |
ENST00000342084.4
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr1_+_19578033 | 0.69 |
ENST00000330263.4
|
MRTO4
|
mRNA turnover 4 homolog (S. cerevisiae) |
| chr5_+_34757309 | 0.68 |
ENST00000397449.1
|
RAI14
|
retinoic acid induced 14 |
| chr16_+_69373323 | 0.67 |
ENST00000254940.5
|
NIP7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr3_+_100428188 | 0.66 |
ENST00000418917.2
ENST00000490574.1 |
TFG
|
TRK-fused gene |
| chr19_+_42746927 | 0.64 |
ENST00000378108.1
|
AC006486.1
|
AC006486.1 |
| chr22_-_24096630 | 0.64 |
ENST00000248948.3
|
VPREB3
|
pre-B lymphocyte 3 |
| chr11_-_104827425 | 0.64 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr6_+_126102292 | 0.62 |
ENST00000368357.3
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr13_-_26452679 | 0.62 |
ENST00000596729.1
|
AL138815.2
|
Uncharacterized protein |
| chr1_+_16767167 | 0.59 |
ENST00000337132.5
|
NECAP2
|
NECAP endocytosis associated 2 |
| chr4_-_70080449 | 0.58 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr4_-_48082192 | 0.57 |
ENST00000507351.1
|
TXK
|
TXK tyrosine kinase |
| chr19_+_52076425 | 0.55 |
ENST00000436511.2
|
ZNF175
|
zinc finger protein 175 |
| chr19_+_17186577 | 0.54 |
ENST00000595618.1
ENST00000594824.1 |
MYO9B
|
myosin IXB |
| chr3_-_48130707 | 0.54 |
ENST00000360240.6
ENST00000383737.4 |
MAP4
|
microtubule-associated protein 4 |
| chr14_+_23654525 | 0.53 |
ENST00000399910.1
ENST00000492621.1 |
C14orf164
|
chromosome 14 open reading frame 164 |
| chr5_-_139937895 | 0.52 |
ENST00000336283.6
|
SRA1
|
steroid receptor RNA activator 1 |
| chr6_+_12007897 | 0.51 |
ENST00000437559.1
|
RP11-456H18.2
|
RP11-456H18.2 |
| chr7_+_123241908 | 0.50 |
ENST00000434204.1
ENST00000437535.1 ENST00000451215.1 |
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr8_+_20054878 | 0.49 |
ENST00000276390.2
ENST00000519667.1 |
ATP6V1B2
|
ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 |
| chr19_+_496454 | 0.48 |
ENST00000346144.4
ENST00000215637.3 ENST00000382683.4 |
MADCAM1
|
mucosal vascular addressin cell adhesion molecule 1 |
| chr9_+_131447342 | 0.47 |
ENST00000409104.3
|
SET
|
SET nuclear oncogene |
| chr2_-_179567206 | 0.46 |
ENST00000414766.1
|
TTN
|
titin |
| chr1_+_158259558 | 0.46 |
ENST00000368170.3
|
CD1C
|
CD1c molecule |
| chr11_+_65627865 | 0.45 |
ENST00000308110.4
|
MUS81
|
MUS81 structure-specific endonuclease subunit |
| chr4_+_70146217 | 0.45 |
ENST00000335568.5
ENST00000511240.1 |
UGT2B28
|
UDP glucuronosyltransferase 2 family, polypeptide B28 |
| chr17_-_41116454 | 0.45 |
ENST00000427569.2
ENST00000430739.1 |
AARSD1
|
alanyl-tRNA synthetase domain containing 1 |
| chr1_+_16767195 | 0.44 |
ENST00000504551.2
ENST00000457722.2 ENST00000406746.1 ENST00000443980.2 |
NECAP2
|
NECAP endocytosis associated 2 |
| chr20_-_1373606 | 0.43 |
ENST00000381715.1
ENST00000439640.2 ENST00000381719.3 |
FKBP1A
|
FK506 binding protein 1A, 12kDa |
| chr1_-_200638964 | 0.43 |
ENST00000367348.3
ENST00000447706.2 ENST00000331314.6 |
DDX59
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 59 |
| chr11_+_111126707 | 0.42 |
ENST00000280325.4
|
C11orf53
|
chromosome 11 open reading frame 53 |
| chr12_+_70219052 | 0.41 |
ENST00000552032.2
ENST00000547771.2 |
MYRFL
|
myelin regulatory factor-like |
| chr15_-_41120896 | 0.41 |
ENST00000299174.5
ENST00000427255.2 |
PPP1R14D
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr2_-_175462456 | 0.40 |
ENST00000409891.1
ENST00000410117.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr4_-_156875003 | 0.39 |
ENST00000433477.3
|
CTSO
|
cathepsin O |
| chr1_+_169337172 | 0.38 |
ENST00000367807.3
ENST00000367808.3 ENST00000329281.2 ENST00000420531.1 |
BLZF1
|
basic leucine zipper nuclear factor 1 |
| chr4_-_170924888 | 0.37 |
ENST00000502832.1
ENST00000393704.3 |
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr5_+_140071011 | 0.37 |
ENST00000230771.3
ENST00000509299.1 ENST00000503873.1 ENST00000435019.2 ENST00000437649.2 ENST00000432671.2 |
HARS2
|
histidyl-tRNA synthetase 2, mitochondrial |
| chr2_-_163099885 | 0.37 |
ENST00000443424.1
|
FAP
|
fibroblast activation protein, alpha |
| chr1_-_173793458 | 0.37 |
ENST00000356198.2
|
CENPL
|
centromere protein L |
| chr17_-_34890665 | 0.37 |
ENST00000586007.1
|
MYO19
|
myosin XIX |
| chrX_-_132231123 | 0.37 |
ENST00000511190.1
|
USP26
|
ubiquitin specific peptidase 26 |
| chr11_-_61687739 | 0.36 |
ENST00000531922.1
ENST00000301773.5 |
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr22_-_43539346 | 0.36 |
ENST00000327555.5
ENST00000290429.6 |
MCAT
|
malonyl CoA:ACP acyltransferase (mitochondrial) |
| chr16_-_69373396 | 0.36 |
ENST00000562595.1
ENST00000562081.1 ENST00000306875.4 |
COG8
|
component of oligomeric golgi complex 8 |
| chr10_+_135204338 | 0.36 |
ENST00000468317.2
|
RP11-108K14.8
|
Mitochondrial GTPase 1 |
| chr10_-_27529486 | 0.36 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr1_+_202431859 | 0.35 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr12_-_50294033 | 0.35 |
ENST00000552669.1
|
FAIM2
|
Fas apoptotic inhibitory molecule 2 |
| chr11_-_66360548 | 0.34 |
ENST00000333861.3
|
CCDC87
|
coiled-coil domain containing 87 |
| chr5_+_78407602 | 0.33 |
ENST00000274353.5
ENST00000524080.1 |
BHMT
|
betaine--homocysteine S-methyltransferase |
| chr11_-_64527425 | 0.33 |
ENST00000377432.3
|
PYGM
|
phosphorylase, glycogen, muscle |
| chrX_+_47053208 | 0.33 |
ENST00000442035.1
ENST00000457753.1 ENST00000335972.6 |
UBA1
|
ubiquitin-like modifier activating enzyme 1 |
| chr1_+_173793777 | 0.33 |
ENST00000239457.5
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr7_+_100466433 | 0.33 |
ENST00000429658.1
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr17_+_60501228 | 0.33 |
ENST00000311506.5
|
METTL2A
|
methyltransferase like 2A |
| chr2_-_163100045 | 0.33 |
ENST00000188790.4
|
FAP
|
fibroblast activation protein, alpha |
| chr3_-_42003479 | 0.33 |
ENST00000420927.1
|
ULK4
|
unc-51 like kinase 4 |
| chr2_-_175462934 | 0.32 |
ENST00000392546.2
ENST00000436221.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr16_-_20367584 | 0.32 |
ENST00000570689.1
|
UMOD
|
uromodulin |
| chr6_+_42584847 | 0.31 |
ENST00000372883.3
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr17_-_2966901 | 0.30 |
ENST00000575751.1
|
OR1D5
|
olfactory receptor, family 1, subfamily D, member 5 |
| chr15_+_81475047 | 0.30 |
ENST00000559388.1
|
IL16
|
interleukin 16 |
| chr9_-_137809718 | 0.30 |
ENST00000371806.3
|
FCN1
|
ficolin (collagen/fibrinogen domain containing) 1 |
| chr19_-_51611623 | 0.30 |
ENST00000421832.2
|
CTU1
|
cytosolic thiouridylase subunit 1 |
| chr12_-_44152551 | 0.29 |
ENST00000416848.2
ENST00000550784.1 ENST00000547156.1 ENST00000549868.1 ENST00000553166.1 ENST00000551923.1 ENST00000431332.3 ENST00000344862.5 |
PUS7L
|
pseudouridylate synthase 7 homolog (S. cerevisiae)-like |
| chr17_+_33914276 | 0.29 |
ENST00000592545.1
ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr4_+_91048706 | 0.28 |
ENST00000509176.1
|
CCSER1
|
coiled-coil serine-rich protein 1 |
| chr2_-_69180083 | 0.28 |
ENST00000328895.4
|
GKN2
|
gastrokine 2 |
| chr22_-_50523760 | 0.27 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr11_-_105892937 | 0.27 |
ENST00000301919.4
ENST00000534458.1 ENST00000530108.1 ENST00000530788.1 |
MSANTD4
|
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
| chr7_+_110731062 | 0.27 |
ENST00000308478.5
ENST00000451085.1 ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chr22_-_24096562 | 0.27 |
ENST00000398465.3
|
VPREB3
|
pre-B lymphocyte 3 |
| chr20_+_13202418 | 0.27 |
ENST00000262487.4
|
ISM1
|
isthmin 1, angiogenesis inhibitor |
| chr8_+_50824233 | 0.27 |
ENST00000522124.1
|
SNTG1
|
syntrophin, gamma 1 |
| chr1_-_173793246 | 0.27 |
ENST00000345664.6
ENST00000367710.3 |
CENPL
|
centromere protein L |
| chr4_-_73434498 | 0.27 |
ENST00000286657.4
|
ADAMTS3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr2_-_179516225 | 0.26 |
ENST00000446966.1
ENST00000426232.1 |
TTN
|
titin |
| chr19_+_52074502 | 0.26 |
ENST00000545217.1
ENST00000262259.2 ENST00000596504.1 |
ZNF175
|
zinc finger protein 175 |
| chr8_-_30706608 | 0.25 |
ENST00000256246.2
|
TEX15
|
testis expressed 15 |
| chr17_-_7216939 | 0.25 |
ENST00000573684.1
|
GPS2
|
G protein pathway suppressor 2 |
| chr17_+_33914460 | 0.24 |
ENST00000537622.2
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr5_-_140070897 | 0.23 |
ENST00000448240.1
ENST00000438307.2 ENST00000415192.2 ENST00000457527.2 ENST00000307633.3 ENST00000507746.1 ENST00000431330.2 |
HARS
|
histidyl-tRNA synthetase |
| chr6_+_10528560 | 0.23 |
ENST00000379597.3
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr6_-_34664612 | 0.23 |
ENST00000374023.3
ENST00000374026.3 |
C6orf106
|
chromosome 6 open reading frame 106 |
| chr12_-_57039739 | 0.23 |
ENST00000552959.1
ENST00000551020.1 ENST00000553007.2 ENST00000552919.1 ENST00000552104.1 ENST00000262030.3 |
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr14_-_30766223 | 0.23 |
ENST00000549360.1
ENST00000508469.2 |
CTD-2251F13.1
|
CTD-2251F13.1 |
| chr7_+_23719749 | 0.22 |
ENST00000409192.3
ENST00000344962.4 ENST00000409653.1 ENST00000409994.3 |
FAM221A
|
family with sequence similarity 221, member A |
| chr4_+_169418195 | 0.22 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr14_-_47812321 | 0.22 |
ENST00000357362.3
ENST00000486952.2 ENST00000426342.1 |
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chr17_-_34890709 | 0.21 |
ENST00000544606.1
|
MYO19
|
myosin XIX |
| chr10_+_18629628 | 0.21 |
ENST00000377329.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr19_+_56111680 | 0.21 |
ENST00000301073.3
|
ZNF524
|
zinc finger protein 524 |
| chr7_-_91509986 | 0.21 |
ENST00000456229.1
ENST00000442961.1 ENST00000406735.2 ENST00000419292.1 ENST00000351870.3 |
MTERF
|
mitochondrial transcription termination factor |
| chr1_+_110163202 | 0.20 |
ENST00000531203.1
ENST00000256578.3 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr17_-_34890732 | 0.20 |
ENST00000268852.9
|
MYO19
|
myosin XIX |
| chr7_+_121513143 | 0.20 |
ENST00000393386.2
|
PTPRZ1
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1 |
| chr3_+_121796697 | 0.20 |
ENST00000482356.1
ENST00000393627.2 |
CD86
|
CD86 molecule |
| chr2_-_25565377 | 0.19 |
ENST00000264709.3
ENST00000406659.3 |
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha |
| chr2_+_12246664 | 0.19 |
ENST00000449986.1
|
AC096559.1
|
AC096559.1 |
| chr16_-_47493041 | 0.19 |
ENST00000565940.2
|
ITFG1
|
integrin alpha FG-GAP repeat containing 1 |
| chr17_-_26903900 | 0.19 |
ENST00000395319.3
ENST00000581807.1 ENST00000584086.1 ENST00000395321.2 |
ALDOC
|
aldolase C, fructose-bisphosphate |
| chr5_+_140071178 | 0.19 |
ENST00000508522.1
ENST00000448069.2 |
HARS2
|
histidyl-tRNA synthetase 2, mitochondrial |
| chr17_-_74049720 | 0.18 |
ENST00000602720.1
|
SRP68
|
signal recognition particle 68kDa |
| chr11_+_27062860 | 0.17 |
ENST00000528583.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr9_+_4839762 | 0.17 |
ENST00000448872.2
ENST00000441844.1 |
RCL1
|
RNA terminal phosphate cyclase-like 1 |
| chr3_-_113956425 | 0.17 |
ENST00000482457.2
|
ZNF80
|
zinc finger protein 80 |
| chr14_-_50778872 | 0.17 |
ENST00000555610.1
ENST00000261699.4 |
L2HGDH
|
L-2-hydroxyglutarate dehydrogenase |
| chr1_+_173793641 | 0.17 |
ENST00000361951.4
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr10_+_32735030 | 0.17 |
ENST00000277657.6
ENST00000362006.5 |
CCDC7
|
coiled-coil domain containing 7 |
| chr5_+_175487692 | 0.16 |
ENST00000510151.1
|
FAM153B
|
family with sequence similarity 153, member B |
| chr2_-_87088995 | 0.16 |
ENST00000393759.2
ENST00000349455.3 ENST00000331469.2 ENST00000431506.2 ENST00000393761.2 ENST00000390655.6 |
CD8B
|
CD8b molecule |
| chr3_+_114012819 | 0.15 |
ENST00000383671.3
|
TIGIT
|
T cell immunoreceptor with Ig and ITIM domains |
| chr5_-_177210399 | 0.15 |
ENST00000510276.1
|
FAM153A
|
family with sequence similarity 153, member A |
| chr17_-_34890759 | 0.15 |
ENST00000431794.3
|
MYO19
|
myosin XIX |
| chr11_+_27062502 | 0.14 |
ENST00000263182.3
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chrX_+_139791917 | 0.14 |
ENST00000607004.1
ENST00000370535.3 |
LINC00632
|
long intergenic non-protein coding RNA 632 |
| chr5_-_169725231 | 0.14 |
ENST00000046794.5
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
| chr14_+_50779029 | 0.14 |
ENST00000245448.6
|
ATP5S
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit s (factor B) |
| chr17_-_39341594 | 0.14 |
ENST00000398472.1
|
KRTAP4-1
|
keratin associated protein 4-1 |
| chr20_-_21494654 | 0.14 |
ENST00000377142.4
|
NKX2-2
|
NK2 homeobox 2 |
| chr9_-_215744 | 0.13 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr13_+_115000521 | 0.13 |
ENST00000252457.5
ENST00000375308.1 |
CDC16
|
cell division cycle 16 |
| chr1_+_158223923 | 0.13 |
ENST00000289429.5
|
CD1A
|
CD1a molecule |
| chr1_-_78444776 | 0.13 |
ENST00000370767.1
ENST00000421641.1 |
FUBP1
|
far upstream element (FUSE) binding protein 1 |
| chr1_-_78444738 | 0.12 |
ENST00000436586.2
ENST00000370768.2 |
FUBP1
|
far upstream element (FUSE) binding protein 1 |
| chr19_+_55385682 | 0.12 |
ENST00000391726.3
|
FCAR
|
Fc fragment of IgA, receptor for |
| chr1_-_108231101 | 0.12 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr22_+_42017987 | 0.12 |
ENST00000405506.1
|
XRCC6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr8_-_95220775 | 0.12 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr1_+_113010056 | 0.12 |
ENST00000369686.5
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
| chr19_-_49843539 | 0.12 |
ENST00000602554.1
ENST00000358234.4 |
CTC-301O7.4
|
CTC-301O7.4 |
| chr1_-_31902614 | 0.12 |
ENST00000596131.1
|
AC114494.1
|
HCG1787699; Uncharacterized protein |
| chr15_+_84841242 | 0.11 |
ENST00000558195.1
|
RP11-182J1.16
|
ubiquitin-conjugating enzyme E2Q family member 2-like |
| chr3_-_46249878 | 0.11 |
ENST00000296140.3
|
CCR1
|
chemokine (C-C motif) receptor 1 |
| chr4_+_186317133 | 0.11 |
ENST00000507753.1
|
ANKRD37
|
ankyrin repeat domain 37 |
| chr7_+_141463897 | 0.11 |
ENST00000247879.2
|
TAS2R3
|
taste receptor, type 2, member 3 |
| chr1_+_20512568 | 0.11 |
ENST00000375099.3
|
UBXN10
|
UBX domain protein 10 |
| chr3_-_81811312 | 0.11 |
ENST00000429644.2
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1 |
| chr11_-_65624415 | 0.11 |
ENST00000524553.1
ENST00000527344.1 |
CFL1
|
cofilin 1 (non-muscle) |
| chr15_-_28344439 | 0.10 |
ENST00000431101.1
ENST00000445578.1 ENST00000353809.5 ENST00000382996.2 ENST00000354638.3 |
OCA2
|
oculocutaneous albinism II |
| chr4_+_17579110 | 0.10 |
ENST00000606142.1
|
LAP3
|
leucine aminopeptidase 3 |
| chr6_-_15586238 | 0.10 |
ENST00000462989.2
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr11_+_118958689 | 0.10 |
ENST00000535253.1
ENST00000392841.1 |
HMBS
|
hydroxymethylbilane synthase |
| chr14_+_22580233 | 0.09 |
ENST00000390454.2
|
TRAV25
|
T cell receptor alpha variable 25 |
| chr3_-_39196049 | 0.09 |
ENST00000514182.1
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1 |
| chr9_+_131452239 | 0.09 |
ENST00000372688.4
ENST00000372686.5 |
SET
|
SET nuclear oncogene |
| chr3_-_157824292 | 0.09 |
ENST00000483851.2
|
SHOX2
|
short stature homeobox 2 |
| chr13_+_115000556 | 0.08 |
ENST00000252458.6
|
CDC16
|
cell division cycle 16 |
| chr19_-_37663332 | 0.08 |
ENST00000392157.2
|
ZNF585A
|
zinc finger protein 585A |
| chr11_-_64684672 | 0.08 |
ENST00000377264.3
ENST00000421419.2 |
ATG2A
|
autophagy related 2A |
| chr12_-_50290839 | 0.07 |
ENST00000552863.1
|
FAIM2
|
Fas apoptotic inhibitory molecule 2 |
| chr1_-_160001737 | 0.07 |
ENST00000368090.2
|
PIGM
|
phosphatidylinositol glycan anchor biosynthesis, class M |
| chr12_+_4829507 | 0.07 |
ENST00000252318.2
|
GALNT8
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 8 (GalNAc-T8) |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.6 | 2.3 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.4 | 2.8 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 | 1.0 | GO:0046210 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) |
| 0.3 | 0.8 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.3 | 0.8 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.2 | 1.4 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.2 | 1.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.2 | 1.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.2 | 0.7 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.2 | 0.5 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.2 | 0.9 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.2 | 1.4 | GO:0001554 | luteolysis(GO:0001554) |
| 0.1 | 0.4 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.7 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 1.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 1.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.5 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 0.3 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.1 | 0.8 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.5 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 0.3 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 0.7 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.6 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 1.0 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.5 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.1 | 0.5 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 1.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 1.0 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 0.3 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.2 | GO:0045404 | negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.1 | 0.3 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.1 | 3.2 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.1 | 0.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.3 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.1 | 0.6 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 0.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.6 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 4.5 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.5 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 1.8 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.2 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:0043324 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.2 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:2000466 | negative regulation of glycogen (starch) synthase activity(GO:2000466) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.4 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.2 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 2.9 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 1.8 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.2 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.9 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.2 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.9 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.1 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.6 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.3 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.3 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.6 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.7 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 0.3 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.3 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.5 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.6 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 1.0 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.9 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.3 | 0.9 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.1 | 0.5 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.5 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.1 | 1.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 2.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 1.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 0.6 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 6.4 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.3 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.8 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 1.0 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 1.6 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.5 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.3 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 1.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 8.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.2 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 1.0 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 1.5 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 1.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.6 | 6.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.5 | 1.4 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.4 | 4.6 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.3 | 1.0 | GO:0047726 | iron-cytochrome-c reductase activity(GO:0047726) |
| 0.3 | 0.8 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.2 | 1.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 0.5 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.2 | 0.9 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) |
| 0.2 | 2.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.4 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.3 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 0.1 | 0.9 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.5 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 1.0 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 1.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.3 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.5 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.1 | 1.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 9.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 0.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.6 | GO:0030883 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.1 | 0.7 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 1.0 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.4 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 1.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.5 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.6 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 1.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.4 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.5 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.3 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 1.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 1.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.9 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 3.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.0 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.9 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 2.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 2.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 4.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.5 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.6 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |