Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
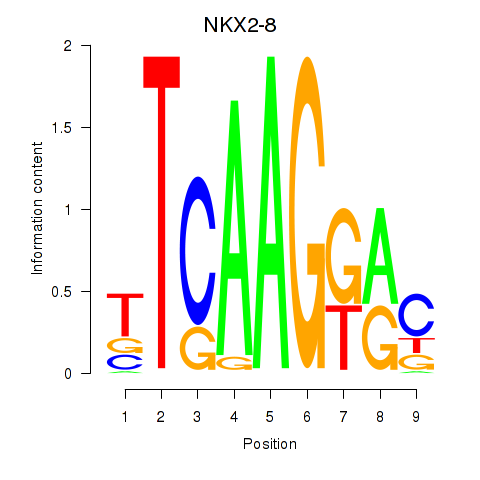
Results for NKX2-8
Z-value: 0.45
Transcription factors associated with NKX2-8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-8
|
ENSG00000136327.6 | NK2 homeobox 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-8 | hg19_v2_chr14_-_37051798_37051831 | -0.33 | 1.1e-01 | Click! |
Activity profile of NKX2-8 motif
Sorted Z-values of NKX2-8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_88872176 | 0.65 |
ENST00000569140.1
|
CDT1
|
chromatin licensing and DNA replication factor 1 |
| chr4_-_70080449 | 0.46 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr1_+_74701062 | 0.31 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr3_+_126423045 | 0.30 |
ENST00000290913.3
ENST00000508789.1 |
CHCHD6
|
coiled-coil-helix-coiled-coil-helix domain containing 6 |
| chr11_+_5410607 | 0.28 |
ENST00000328611.3
|
OR51M1
|
olfactory receptor, family 51, subfamily M, member 1 |
| chr3_-_116163830 | 0.24 |
ENST00000333617.4
|
LSAMP
|
limbic system-associated membrane protein |
| chr2_-_29297127 | 0.23 |
ENST00000331664.5
|
C2orf71
|
chromosome 2 open reading frame 71 |
| chr5_+_3596168 | 0.22 |
ENST00000302006.3
|
IRX1
|
iroquois homeobox 1 |
| chr3_+_111717600 | 0.22 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr11_-_107590383 | 0.22 |
ENST00000525934.1
ENST00000531293.1 |
SLN
|
sarcolipin |
| chr18_-_44336998 | 0.21 |
ENST00000315087.7
|
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr3_+_111717511 | 0.21 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr20_+_31619454 | 0.19 |
ENST00000349552.1
|
BPIFB6
|
BPI fold containing family B, member 6 |
| chr19_-_38720354 | 0.18 |
ENST00000416611.1
|
DPF1
|
D4, zinc and double PHD fingers family 1 |
| chr15_+_93749295 | 0.17 |
ENST00000599897.1
|
AC112693.2
|
AC112693.2 |
| chrX_-_69479654 | 0.17 |
ENST00000374519.2
|
P2RY4
|
pyrimidinergic receptor P2Y, G-protein coupled, 4 |
| chr14_-_107131560 | 0.16 |
ENST00000390632.2
|
IGHV3-66
|
immunoglobulin heavy variable 3-66 |
| chr6_-_32145861 | 0.16 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr12_-_12837423 | 0.15 |
ENST00000540510.1
|
GPR19
|
G protein-coupled receptor 19 |
| chr12_-_75603482 | 0.15 |
ENST00000341669.3
ENST00000298972.1 ENST00000350228.2 |
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chrX_-_70326455 | 0.15 |
ENST00000374251.5
|
CXorf65
|
chromosome X open reading frame 65 |
| chr14_-_107049312 | 0.15 |
ENST00000390627.2
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr12_-_75603202 | 0.15 |
ENST00000393288.2
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr11_-_72353451 | 0.15 |
ENST00000376450.3
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated |
| chr12_-_75603643 | 0.15 |
ENST00000549446.1
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chrX_-_110655306 | 0.15 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr7_+_129847688 | 0.15 |
ENST00000297819.3
|
SSMEM1
|
serine-rich single-pass membrane protein 1 |
| chr16_-_66952742 | 0.14 |
ENST00000565235.2
ENST00000568632.1 ENST00000565796.1 |
CDH16
|
cadherin 16, KSP-cadherin |
| chrX_-_151143140 | 0.14 |
ENST00000393914.3
ENST00000370328.3 ENST00000370325.1 |
GABRE
|
gamma-aminobutyric acid (GABA) A receptor, epsilon |
| chr17_+_73642486 | 0.14 |
ENST00000579469.1
|
SMIM6
|
small integral membrane protein 6 |
| chr12_+_25205666 | 0.14 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr3_+_67048721 | 0.14 |
ENST00000295568.4
ENST00000484414.1 ENST00000460576.1 ENST00000417314.2 |
KBTBD8
|
kelch repeat and BTB (POZ) domain containing 8 |
| chr10_-_135171178 | 0.14 |
ENST00000368551.1
|
FUOM
|
fucose mutarotase |
| chr4_+_76481258 | 0.14 |
ENST00000311623.4
ENST00000435974.2 |
C4orf26
|
chromosome 4 open reading frame 26 |
| chr12_-_75603236 | 0.13 |
ENST00000540018.1
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr11_-_5364809 | 0.13 |
ENST00000300773.2
|
OR51B5
|
olfactory receptor, family 51, subfamily B, member 5 |
| chr16_-_66952779 | 0.13 |
ENST00000570262.1
ENST00000394055.3 ENST00000299752.4 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr3_-_58613323 | 0.13 |
ENST00000474531.1
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107, member A |
| chr17_+_73642315 | 0.13 |
ENST00000556126.2
|
SMIM6
|
small integral membrane protein 6 |
| chr4_+_184826418 | 0.12 |
ENST00000308497.4
ENST00000438269.1 |
STOX2
|
storkhead box 2 |
| chrX_-_110654147 | 0.12 |
ENST00000358070.4
|
DCX
|
doublecortin |
| chr17_+_76227391 | 0.12 |
ENST00000586400.1
ENST00000421688.1 ENST00000374946.3 |
TMEM235
|
transmembrane protein 235 |
| chr5_-_176056974 | 0.12 |
ENST00000510387.1
ENST00000506696.1 |
SNCB
|
synuclein, beta |
| chr8_+_23430157 | 0.12 |
ENST00000399967.3
|
FP15737
|
FP15737 |
| chr21_+_33671264 | 0.12 |
ENST00000339944.4
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr12_+_60058458 | 0.11 |
ENST00000548610.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr5_-_101834712 | 0.11 |
ENST00000506729.1
ENST00000389019.3 ENST00000379810.1 |
SLCO6A1
|
solute carrier organic anion transporter family, member 6A1 |
| chr1_+_16348525 | 0.11 |
ENST00000331433.4
|
CLCNKA
|
chloride channel, voltage-sensitive Ka |
| chr4_+_37245799 | 0.11 |
ENST00000309447.5
|
KIAA1239
|
KIAA1239 |
| chr5_-_101834617 | 0.11 |
ENST00000513675.1
ENST00000379807.3 |
SLCO6A1
|
solute carrier organic anion transporter family, member 6A1 |
| chr12_+_66218903 | 0.11 |
ENST00000393577.3
|
HMGA2
|
high mobility group AT-hook 2 |
| chr1_-_155006224 | 0.11 |
ENST00000368424.3
|
DCST2
|
DC-STAMP domain containing 2 |
| chr17_+_73472575 | 0.11 |
ENST00000375248.5
|
KIAA0195
|
KIAA0195 |
| chr14_-_106552755 | 0.10 |
ENST00000390600.2
|
IGHV3-9
|
immunoglobulin heavy variable 3-9 |
| chr3_+_108855558 | 0.10 |
ENST00000467240.1
ENST00000477643.1 ENST00000479039.1 ENST00000593799.1 |
RP11-59E19.1
|
RP11-59E19.1 |
| chr2_-_159313214 | 0.10 |
ENST00000409889.1
ENST00000283233.5 ENST00000536771.1 |
CCDC148
|
coiled-coil domain containing 148 |
| chr16_+_32063311 | 0.10 |
ENST00000426099.1
|
AC142381.1
|
AC142381.1 |
| chr3_-_196242233 | 0.10 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr19_+_34856141 | 0.10 |
ENST00000586425.1
|
GPI
|
glucose-6-phosphate isomerase |
| chr12_+_121416489 | 0.10 |
ENST00000541395.1
ENST00000544413.1 |
HNF1A
|
HNF1 homeobox A |
| chr3_-_114790179 | 0.10 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr3_+_173302222 | 0.10 |
ENST00000361589.4
|
NLGN1
|
neuroligin 1 |
| chr2_-_46385 | 0.10 |
ENST00000327669.4
|
FAM110C
|
family with sequence similarity 110, member C |
| chr1_+_180165672 | 0.09 |
ENST00000443059.1
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr14_-_107199464 | 0.09 |
ENST00000433072.2
|
IGHV3-72
|
immunoglobulin heavy variable 3-72 |
| chr14_-_24732368 | 0.09 |
ENST00000544573.1
|
TGM1
|
transglutaminase 1 |
| chr11_+_35211429 | 0.09 |
ENST00000525688.1
ENST00000278385.6 ENST00000533222.1 |
CD44
|
CD44 molecule (Indian blood group) |
| chr14_-_58894223 | 0.09 |
ENST00000555593.1
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr15_-_28419569 | 0.09 |
ENST00000569772.1
|
HERC2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr14_-_58894332 | 0.09 |
ENST00000395159.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr16_-_18468926 | 0.09 |
ENST00000545114.1
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr12_-_49504655 | 0.09 |
ENST00000551782.1
ENST00000267102.8 |
LMBR1L
|
limb development membrane protein 1-like |
| chr12_-_49504623 | 0.09 |
ENST00000550137.1
ENST00000547382.1 |
LMBR1L
|
limb development membrane protein 1-like |
| chr10_+_71078595 | 0.09 |
ENST00000359426.6
|
HK1
|
hexokinase 1 |
| chr6_-_32806506 | 0.09 |
ENST00000374897.2
ENST00000452392.2 |
TAP2
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) Uncharacterized protein |
| chr3_-_178984759 | 0.09 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr22_+_38453207 | 0.08 |
ENST00000404072.3
ENST00000424694.1 |
PICK1
|
protein interacting with PRKCA 1 |
| chr15_-_56757329 | 0.08 |
ENST00000260453.3
|
MNS1
|
meiosis-specific nuclear structural 1 |
| chr12_+_66218598 | 0.08 |
ENST00000541363.1
|
HMGA2
|
high mobility group AT-hook 2 |
| chr17_-_79623597 | 0.08 |
ENST00000574024.1
ENST00000331056.5 |
PDE6G
|
phosphodiesterase 6G, cGMP-specific, rod, gamma |
| chr11_-_133826852 | 0.08 |
ENST00000533871.2
ENST00000321016.8 |
IGSF9B
|
immunoglobulin superfamily, member 9B |
| chr14_-_106586656 | 0.08 |
ENST00000390602.2
|
IGHV3-13
|
immunoglobulin heavy variable 3-13 |
| chr19_-_44259136 | 0.08 |
ENST00000270066.6
|
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr6_+_125524785 | 0.08 |
ENST00000392482.2
|
TPD52L1
|
tumor protein D52-like 1 |
| chr8_+_67405794 | 0.08 |
ENST00000522977.1
ENST00000480005.1 |
C8orf46
|
chromosome 8 open reading frame 46 |
| chr6_-_55740352 | 0.08 |
ENST00000370830.3
|
BMP5
|
bone morphogenetic protein 5 |
| chr3_+_63953415 | 0.08 |
ENST00000484332.1
|
ATXN7
|
ataxin 7 |
| chr22_+_29834572 | 0.08 |
ENST00000354373.2
|
RFPL1
|
ret finger protein-like 1 |
| chr1_+_113010056 | 0.08 |
ENST00000369686.5
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
| chr13_+_108921977 | 0.08 |
ENST00000430559.1
ENST00000375887.4 |
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr7_-_156803329 | 0.08 |
ENST00000252971.6
|
MNX1
|
motor neuron and pancreas homeobox 1 |
| chr16_-_33647696 | 0.07 |
ENST00000558425.1
ENST00000569103.2 |
RP11-812E19.9
|
Uncharacterized protein |
| chr14_-_106068065 | 0.07 |
ENST00000390541.2
|
IGHE
|
immunoglobulin heavy constant epsilon |
| chr3_-_53164423 | 0.07 |
ENST00000467048.1
ENST00000394738.3 ENST00000296292.3 |
RFT1
|
RFT1 homolog (S. cerevisiae) |
| chr6_+_69942298 | 0.07 |
ENST00000238918.8
|
BAI3
|
brain-specific angiogenesis inhibitor 3 |
| chr6_-_137113604 | 0.07 |
ENST00000359015.4
|
MAP3K5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr14_-_106994333 | 0.07 |
ENST00000390624.2
|
IGHV3-48
|
immunoglobulin heavy variable 3-48 |
| chrX_-_63450480 | 0.07 |
ENST00000362002.2
|
ASB12
|
ankyrin repeat and SOCS box containing 12 |
| chr9_+_6328360 | 0.07 |
ENST00000381428.1
ENST00000314556.3 |
TPD52L3
|
tumor protein D52-like 3 |
| chr16_+_88493879 | 0.07 |
ENST00000565624.1
ENST00000437464.1 |
ZNF469
|
zinc finger protein 469 |
| chr13_-_103346854 | 0.07 |
ENST00000267273.6
|
METTL21C
|
methyltransferase like 21C |
| chr1_+_173793777 | 0.07 |
ENST00000239457.5
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chrX_-_64196307 | 0.07 |
ENST00000545618.1
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr2_+_207024306 | 0.07 |
ENST00000236957.5
ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr1_-_104238912 | 0.07 |
ENST00000330330.5
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chrX_-_64196376 | 0.07 |
ENST00000447788.2
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr20_-_48530230 | 0.07 |
ENST00000422556.1
|
SPATA2
|
spermatogenesis associated 2 |
| chr12_-_11548496 | 0.07 |
ENST00000389362.4
ENST00000565533.1 ENST00000546254.1 |
PRB2
PRB1
|
proline-rich protein BstNI subfamily 2 proline-rich protein BstNI subfamily 1 |
| chr15_+_97326659 | 0.07 |
ENST00000558553.1
|
SPATA8
|
spermatogenesis associated 8 |
| chr12_+_25205446 | 0.07 |
ENST00000557489.1
ENST00000354454.3 ENST00000536173.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr17_-_8198636 | 0.07 |
ENST00000577745.1
ENST00000579192.1 ENST00000396278.1 |
SLC25A35
|
solute carrier family 25, member 35 |
| chr10_+_118305435 | 0.07 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase |
| chr7_+_30634297 | 0.07 |
ENST00000389266.3
|
GARS
|
glycyl-tRNA synthetase |
| chr5_+_149737202 | 0.07 |
ENST00000451292.1
ENST00000377797.3 ENST00000445265.2 ENST00000323668.7 ENST00000439160.2 ENST00000394269.3 ENST00000427724.2 ENST00000504761.2 ENST00000513346.1 ENST00000515516.1 |
TCOF1
|
Treacher Collins-Franceschetti syndrome 1 |
| chr6_-_133084580 | 0.07 |
ENST00000525270.1
ENST00000530536.1 ENST00000524919.1 |
VNN2
|
vanin 2 |
| chr3_-_47950745 | 0.07 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr14_-_106573756 | 0.07 |
ENST00000390601.2
|
IGHV3-11
|
immunoglobulin heavy variable 3-11 (gene/pseudogene) |
| chr16_+_33006369 | 0.06 |
ENST00000425181.3
|
IGHV3OR16-10
|
immunoglobulin heavy variable 3/OR16-10 (non-functional) |
| chr8_+_128426535 | 0.06 |
ENST00000465342.2
|
POU5F1B
|
POU class 5 homeobox 1B |
| chr4_-_69111401 | 0.06 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr10_-_118032697 | 0.06 |
ENST00000439649.3
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr18_-_24722995 | 0.06 |
ENST00000581714.1
|
CHST9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
| chr15_-_99057551 | 0.06 |
ENST00000558256.1
|
FAM169B
|
family with sequence similarity 169, member B |
| chrX_+_144908928 | 0.06 |
ENST00000408967.2
|
TMEM257
|
transmembrane protein 257 |
| chr8_+_125551338 | 0.06 |
ENST00000276689.3
ENST00000518008.1 ENST00000522532.1 ENST00000517367.1 |
NDUFB9
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9, 22kDa |
| chr9_-_21166659 | 0.06 |
ENST00000380225.1
|
IFNA21
|
interferon, alpha 21 |
| chr16_-_3350614 | 0.06 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr6_+_52051171 | 0.06 |
ENST00000340057.1
|
IL17A
|
interleukin 17A |
| chr7_+_75028199 | 0.06 |
ENST00000437796.1
|
TRIM73
|
tripartite motif containing 73 |
| chr14_+_22670455 | 0.06 |
ENST00000390460.1
|
TRAV26-2
|
T cell receptor alpha variable 26-2 |
| chr12_+_25205568 | 0.06 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr6_-_39290744 | 0.06 |
ENST00000507712.1
|
KCNK16
|
potassium channel, subfamily K, member 16 |
| chr15_-_35047166 | 0.06 |
ENST00000290374.4
|
GJD2
|
gap junction protein, delta 2, 36kDa |
| chrX_-_64196351 | 0.06 |
ENST00000374839.3
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr13_+_28712614 | 0.06 |
ENST00000380958.3
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr16_+_67034456 | 0.06 |
ENST00000540579.1
|
CES4A
|
carboxylesterase 4A |
| chr22_-_50523760 | 0.06 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr2_+_79252822 | 0.06 |
ENST00000272324.5
|
REG3G
|
regenerating islet-derived 3 gamma |
| chr16_+_33629600 | 0.05 |
ENST00000562905.2
|
IGHV3OR16-13
|
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
| chr17_+_46184911 | 0.05 |
ENST00000580219.1
ENST00000452859.2 ENST00000393405.2 ENST00000439357.2 ENST00000359238.2 |
SNX11
|
sorting nexin 11 |
| chr11_-_118095801 | 0.05 |
ENST00000356289.5
|
AMICA1
|
adhesion molecule, interacts with CXADR antigen 1 |
| chr22_-_32341336 | 0.05 |
ENST00000248984.3
|
C22orf24
|
chromosome 22 open reading frame 24 |
| chr14_+_58765103 | 0.05 |
ENST00000355431.3
ENST00000348476.3 ENST00000395168.3 |
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr14_+_22356029 | 0.05 |
ENST00000390437.2
|
TRAV12-2
|
T cell receptor alpha variable 12-2 |
| chr19_+_51630287 | 0.05 |
ENST00000599948.1
|
SIGLEC9
|
sialic acid binding Ig-like lectin 9 |
| chr11_-_10715117 | 0.05 |
ENST00000527509.2
|
MRVI1
|
murine retrovirus integration site 1 homolog |
| chr16_+_67034466 | 0.05 |
ENST00000535696.1
|
CES4A
|
carboxylesterase 4A |
| chr14_+_58894103 | 0.05 |
ENST00000354386.6
ENST00000556134.1 |
KIAA0586
|
KIAA0586 |
| chr11_-_104827425 | 0.05 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr16_-_3767506 | 0.05 |
ENST00000538171.1
|
TRAP1
|
TNF receptor-associated protein 1 |
| chr16_+_23194033 | 0.05 |
ENST00000300061.2
|
SCNN1G
|
sodium channel, non-voltage-gated 1, gamma subunit |
| chr17_+_76210267 | 0.05 |
ENST00000301633.4
ENST00000350051.3 ENST00000374948.2 ENST00000590449.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr14_+_24540046 | 0.05 |
ENST00000397016.2
ENST00000537691.1 ENST00000560356.1 ENST00000558450.1 |
CPNE6
|
copine VI (neuronal) |
| chr14_+_58894706 | 0.05 |
ENST00000261244.5
|
KIAA0586
|
KIAA0586 |
| chr15_+_40453204 | 0.05 |
ENST00000287598.6
ENST00000412359.3 |
BUB1B
|
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr5_+_68665608 | 0.05 |
ENST00000509734.1
ENST00000354868.5 ENST00000521422.1 ENST00000354312.3 ENST00000345306.6 |
RAD17
|
RAD17 homolog (S. pombe) |
| chr8_-_145018905 | 0.05 |
ENST00000398774.2
|
PLEC
|
plectin |
| chr16_+_33020496 | 0.05 |
ENST00000565407.2
|
IGHV3OR16-8
|
immunoglobulin heavy variable 3/OR16-8 (non-functional) |
| chr16_-_3767551 | 0.05 |
ENST00000246957.5
|
TRAP1
|
TNF receptor-associated protein 1 |
| chr11_-_19223523 | 0.05 |
ENST00000265968.3
|
CSRP3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr18_+_43304092 | 0.05 |
ENST00000321925.4
ENST00000587601.1 |
SLC14A1
|
solute carrier family 14 (urea transporter), member 1 (Kidd blood group) |
| chr5_-_78808617 | 0.04 |
ENST00000282260.6
ENST00000508576.1 ENST00000535690.1 |
HOMER1
|
homer homolog 1 (Drosophila) |
| chr2_+_79252804 | 0.04 |
ENST00000393897.2
|
REG3G
|
regenerating islet-derived 3 gamma |
| chr20_+_47835884 | 0.04 |
ENST00000371764.4
|
DDX27
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 |
| chr1_+_32671236 | 0.04 |
ENST00000537469.1
ENST00000291358.6 |
IQCC
|
IQ motif containing C |
| chr14_-_106622419 | 0.04 |
ENST00000390604.2
|
IGHV3-16
|
immunoglobulin heavy variable 3-16 (non-functional) |
| chr15_-_70387120 | 0.04 |
ENST00000539550.1
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr3_-_53290016 | 0.04 |
ENST00000423525.2
ENST00000423516.1 ENST00000296289.6 ENST00000462138.1 |
TKT
|
transketolase |
| chr11_+_64808675 | 0.04 |
ENST00000529996.1
|
SAC3D1
|
SAC3 domain containing 1 |
| chr9_+_131873659 | 0.04 |
ENST00000452489.2
ENST00000347048.4 ENST00000357197.4 ENST00000445241.1 ENST00000355007.3 ENST00000414331.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr4_+_71200681 | 0.04 |
ENST00000273936.5
|
CABS1
|
calcium-binding protein, spermatid-specific 1 |
| chr16_+_8807419 | 0.04 |
ENST00000565016.1
ENST00000567812.1 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr3_-_187455680 | 0.04 |
ENST00000438077.1
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr7_+_100318423 | 0.04 |
ENST00000252723.2
|
EPO
|
erythropoietin |
| chr11_-_114466471 | 0.04 |
ENST00000424261.2
|
NXPE4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr7_-_95025661 | 0.04 |
ENST00000542556.1
ENST00000265627.5 ENST00000427422.1 ENST00000451904.1 |
PON1
PON3
|
paraoxonase 1 paraoxonase 3 |
| chr12_-_371994 | 0.04 |
ENST00000343164.4
ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr14_-_106845789 | 0.04 |
ENST00000390617.2
|
IGHV3-35
|
immunoglobulin heavy variable 3-35 (non-functional) |
| chr22_+_21213771 | 0.04 |
ENST00000439214.1
|
SNAP29
|
synaptosomal-associated protein, 29kDa |
| chr3_+_52813932 | 0.04 |
ENST00000537050.1
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr4_-_119274121 | 0.04 |
ENST00000296498.3
|
PRSS12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chrX_-_109590174 | 0.03 |
ENST00000372054.1
|
GNG5P2
|
guanine nucleotide binding protein (G protein), gamma 5 pseudogene 2 |
| chrY_-_25345070 | 0.03 |
ENST00000382510.4
ENST00000426000.2 ENST00000540248.1 ENST00000405239.1 |
DAZ1
|
deleted in azoospermia 1 |
| chr22_+_32754139 | 0.03 |
ENST00000382088.3
|
RFPL3
|
ret finger protein-like 3 |
| chr11_+_27062272 | 0.03 |
ENST00000529202.1
ENST00000533566.1 |
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr4_-_46126093 | 0.03 |
ENST00000295452.4
|
GABRG1
|
gamma-aminobutyric acid (GABA) A receptor, gamma 1 |
| chr11_-_114466477 | 0.03 |
ENST00000375478.3
|
NXPE4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr3_+_47021168 | 0.03 |
ENST00000450053.3
ENST00000292309.5 ENST00000383740.2 |
NBEAL2
|
neurobeachin-like 2 |
| chr8_-_125551278 | 0.03 |
ENST00000519232.1
ENST00000523888.1 ENST00000522810.1 ENST00000519548.1 ENST00000517678.1 ENST00000605953.1 ENST00000276692.6 |
TATDN1
|
TatD DNase domain containing 1 |
| chr2_+_204801471 | 0.03 |
ENST00000316386.6
ENST00000435193.1 |
ICOS
|
inducible T-cell co-stimulator |
| chr12_+_66218212 | 0.03 |
ENST00000393578.3
ENST00000425208.2 ENST00000536545.1 ENST00000354636.3 |
HMGA2
|
high mobility group AT-hook 2 |
| chr3_-_114343039 | 0.03 |
ENST00000481632.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr13_-_46425865 | 0.03 |
ENST00000400405.2
|
SIAH3
|
siah E3 ubiquitin protein ligase family member 3 |
| chr6_-_160679905 | 0.03 |
ENST00000366953.3
|
SLC22A2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chrX_-_48858667 | 0.03 |
ENST00000376423.4
ENST00000376441.1 |
GRIPAP1
|
GRIP1 associated protein 1 |
| chr2_-_55277436 | 0.03 |
ENST00000354474.6
|
RTN4
|
reticulon 4 |
| chr12_+_51442101 | 0.03 |
ENST00000550929.1
ENST00000262055.4 ENST00000550442.1 ENST00000549340.1 ENST00000548209.1 ENST00000548251.1 ENST00000550814.1 ENST00000547660.1 ENST00000380123.2 ENST00000548401.1 ENST00000418425.2 ENST00000547008.1 ENST00000552739.1 |
LETMD1
|
LETM1 domain containing 1 |
| chr17_+_76210367 | 0.02 |
ENST00000592734.1
ENST00000587746.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chrX_-_15332665 | 0.02 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr2_-_55277512 | 0.02 |
ENST00000402434.2
|
RTN4
|
reticulon 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.1 | 0.6 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 | 0.2 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.1 | 0.2 | GO:0003131 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.1 | 0.2 | GO:0072268 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.1 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.0 | 0.1 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.0 | 0.1 | GO:0035565 | regulation of pronephros size(GO:0035565) renal glucose absorption(GO:0035623) |
| 0.0 | 0.1 | GO:0099545 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.1 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.0 | 0.1 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0015960 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.0 | 0.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.1 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.0 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.3 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.0 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.0 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.1 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.2 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.0 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.0 | GO:1904450 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.0 | 0.5 | GO:0008210 | estrogen metabolic process(GO:0008210) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.0 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.0 | 0.1 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.1 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.0 | 0.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.2 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.0 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.1 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.0 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.0 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |