Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
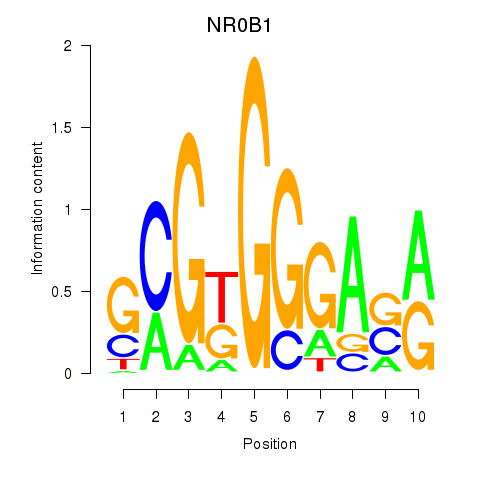
Results for NR0B1
Z-value: 0.73
Transcription factors associated with NR0B1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR0B1
|
ENSG00000169297.6 | nuclear receptor subfamily 0 group B member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR0B1 | hg19_v2_chrX_-_30327495_30327509, hg19_v2_chrX_-_30326445_30326605 | 0.23 | 2.7e-01 | Click! |
Activity profile of NR0B1 motif
Sorted Z-values of NR0B1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_71320557 | 2.73 |
ENST00000541509.1
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta |
| chr7_+_89841144 | 1.97 |
ENST00000394622.2
ENST00000394632.1 ENST00000426158.1 ENST00000394621.2 ENST00000402625.2 |
STEAP2
|
STEAP family member 2, metalloreductase |
| chr7_+_89841024 | 1.94 |
ENST00000394626.1
|
STEAP2
|
STEAP family member 2, metalloreductase |
| chr1_+_212458834 | 1.85 |
ENST00000261461.2
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr22_+_51112800 | 1.80 |
ENST00000414786.2
|
SHANK3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr18_-_22932080 | 1.78 |
ENST00000584787.1
ENST00000361524.3 ENST00000538137.2 |
ZNF521
|
zinc finger protein 521 |
| chr15_-_52970820 | 1.77 |
ENST00000261844.7
ENST00000399202.4 ENST00000562135.1 |
FAM214A
|
family with sequence similarity 214, member A |
| chr9_+_128509624 | 1.66 |
ENST00000342287.5
ENST00000373487.4 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr4_-_90758227 | 1.63 |
ENST00000506691.1
ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr4_-_90758118 | 1.45 |
ENST00000420646.2
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr2_-_152684977 | 1.39 |
ENST00000428992.2
ENST00000295087.8 |
ARL5A
|
ADP-ribosylation factor-like 5A |
| chr12_+_56661461 | 1.28 |
ENST00000546544.1
ENST00000553234.1 |
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae) |
| chr13_-_52027134 | 1.19 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr5_+_61708582 | 1.15 |
ENST00000325324.6
|
IPO11
|
importin 11 |
| chr11_-_2160180 | 1.14 |
ENST00000381406.4
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr1_-_147245484 | 1.11 |
ENST00000271348.2
|
GJA5
|
gap junction protein, alpha 5, 40kDa |
| chr17_+_42634844 | 1.09 |
ENST00000315323.3
|
FZD2
|
frizzled family receptor 2 |
| chr1_+_211432700 | 1.03 |
ENST00000452621.2
|
RCOR3
|
REST corepressor 3 |
| chr4_-_135122903 | 1.00 |
ENST00000421491.3
ENST00000529122.2 |
PABPC4L
|
poly(A) binding protein, cytoplasmic 4-like |
| chr16_+_4845379 | 1.00 |
ENST00000588606.1
ENST00000586005.1 |
SMIM22
|
small integral membrane protein 22 |
| chr1_+_110754094 | 0.96 |
ENST00000369787.3
ENST00000413138.3 ENST00000438661.2 |
KCNC4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4 |
| chr3_-_169381183 | 0.96 |
ENST00000494292.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr14_-_23822061 | 0.95 |
ENST00000397260.3
|
SLC22A17
|
solute carrier family 22, member 17 |
| chr7_+_2559399 | 0.94 |
ENST00000222725.5
ENST00000359574.3 |
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr2_+_170683979 | 0.88 |
ENST00000418381.1
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr6_-_111804905 | 0.87 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr9_+_128509663 | 0.84 |
ENST00000373489.5
ENST00000373483.2 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr19_+_35842445 | 0.83 |
ENST00000246553.2
|
FFAR1
|
free fatty acid receptor 1 |
| chr6_-_111804393 | 0.82 |
ENST00000368802.3
ENST00000368805.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr16_+_89894875 | 0.81 |
ENST00000393062.2
|
SPIRE2
|
spire-type actin nucleation factor 2 |
| chr14_-_23822080 | 0.81 |
ENST00000397267.1
ENST00000354772.3 |
SLC22A17
|
solute carrier family 22, member 17 |
| chr19_-_2050852 | 0.81 |
ENST00000541165.1
ENST00000591601.1 |
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr4_+_88928777 | 0.80 |
ENST00000237596.2
|
PKD2
|
polycystic kidney disease 2 (autosomal dominant) |
| chr12_+_57482665 | 0.79 |
ENST00000300131.3
|
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr20_+_306177 | 0.79 |
ENST00000544632.1
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr15_-_82338460 | 0.79 |
ENST00000558133.1
ENST00000329713.4 |
MEX3B
|
mex-3 RNA binding family member B |
| chr2_+_170683942 | 0.78 |
ENST00000272793.5
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chrX_-_119695279 | 0.77 |
ENST00000336592.6
|
CUL4B
|
cullin 4B |
| chr12_+_56661033 | 0.77 |
ENST00000433805.2
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae) |
| chr13_-_110438914 | 0.76 |
ENST00000375856.3
|
IRS2
|
insulin receptor substrate 2 |
| chr2_+_159313452 | 0.76 |
ENST00000389757.3
ENST00000389759.3 |
PKP4
|
plakophilin 4 |
| chr15_-_65067773 | 0.76 |
ENST00000300069.4
|
RBPMS2
|
RNA binding protein with multiple splicing 2 |
| chr5_+_139028510 | 0.73 |
ENST00000502336.1
ENST00000520967.1 ENST00000511048.1 |
CXXC5
|
CXXC finger protein 5 |
| chr5_-_172198190 | 0.73 |
ENST00000239223.3
|
DUSP1
|
dual specificity phosphatase 1 |
| chr20_+_306221 | 0.73 |
ENST00000342665.2
|
SOX12
|
SRY (sex determining region Y)-box 12 |
| chr5_-_107717058 | 0.72 |
ENST00000359660.5
|
FBXL17
|
F-box and leucine-rich repeat protein 17 |
| chr9_+_71320596 | 0.71 |
ENST00000265382.3
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta |
| chrX_+_9754461 | 0.71 |
ENST00000380913.3
|
SHROOM2
|
shroom family member 2 |
| chrX_+_130192318 | 0.70 |
ENST00000370922.1
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr17_+_81037473 | 0.68 |
ENST00000320095.7
|
METRNL
|
meteorin, glial cell differentiation regulator-like |
| chr6_+_17393888 | 0.68 |
ENST00000493172.1
ENST00000465994.1 |
CAP2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr5_+_175298573 | 0.67 |
ENST00000512824.1
|
CPLX2
|
complexin 2 |
| chr20_-_35807970 | 0.66 |
ENST00000400440.2
ENST00000421643.1 |
MROH8
|
maestro heat-like repeat family member 8 |
| chr22_-_36903101 | 0.65 |
ENST00000397224.4
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr6_+_17393839 | 0.64 |
ENST00000489374.1
ENST00000378990.2 |
CAP2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr11_-_63536113 | 0.63 |
ENST00000433688.1
ENST00000546282.2 |
C11orf95
RP11-466C23.4
|
chromosome 11 open reading frame 95 RP11-466C23.4 |
| chr6_-_112575687 | 0.63 |
ENST00000521398.1
ENST00000424408.2 ENST00000243219.3 |
LAMA4
|
laminin, alpha 4 |
| chr14_+_57046530 | 0.62 |
ENST00000536419.1
ENST00000538838.1 |
TMEM260
|
transmembrane protein 260 |
| chr19_-_14201776 | 0.61 |
ENST00000269724.5
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr1_+_211433275 | 0.59 |
ENST00000367005.4
|
RCOR3
|
REST corepressor 3 |
| chr1_+_153700518 | 0.58 |
ENST00000318967.2
ENST00000456435.1 ENST00000435409.2 |
INTS3
|
integrator complex subunit 3 |
| chrX_-_128788914 | 0.58 |
ENST00000429967.1
ENST00000307484.6 |
APLN
|
apelin |
| chr8_+_100025476 | 0.57 |
ENST00000355155.1
ENST00000357162.2 ENST00000358544.2 ENST00000395996.1 ENST00000441350.2 |
VPS13B
|
vacuolar protein sorting 13 homolog B (yeast) |
| chr1_-_236030216 | 0.56 |
ENST00000389794.3
ENST00000389793.2 |
LYST
|
lysosomal trafficking regulator |
| chr6_-_31865452 | 0.56 |
ENST00000375530.4
ENST00000375537.4 |
EHMT2
|
euchromatic histone-lysine N-methyltransferase 2 |
| chrX_-_112084043 | 0.56 |
ENST00000304758.1
|
AMOT
|
angiomotin |
| chr14_+_57046500 | 0.56 |
ENST00000261556.6
|
TMEM260
|
transmembrane protein 260 |
| chr1_-_171711177 | 0.54 |
ENST00000415773.1
ENST00000367740.2 |
VAMP4
|
vesicle-associated membrane protein 4 |
| chr10_-_123687431 | 0.54 |
ENST00000423243.1
|
ATE1
|
arginyltransferase 1 |
| chr4_+_145567173 | 0.54 |
ENST00000296575.3
|
HHIP
|
hedgehog interacting protein |
| chr1_+_211432593 | 0.53 |
ENST00000367006.4
|
RCOR3
|
REST corepressor 3 |
| chr16_-_2246436 | 0.53 |
ENST00000343516.6
|
CASKIN1
|
CASK interacting protein 1 |
| chr1_+_206138884 | 0.52 |
ENST00000341209.5
ENST00000607379.1 |
FAM72A
|
family with sequence similarity 72, member A |
| chr8_-_100025238 | 0.52 |
ENST00000521696.1
|
RP11-410L14.2
|
RP11-410L14.2 |
| chr6_-_112575838 | 0.52 |
ENST00000455073.1
|
LAMA4
|
laminin, alpha 4 |
| chr7_+_94139105 | 0.52 |
ENST00000297273.4
|
CASD1
|
CAS1 domain containing 1 |
| chr19_-_2051223 | 0.51 |
ENST00000309340.7
ENST00000589534.1 ENST00000250896.3 ENST00000589509.1 |
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr1_+_26146397 | 0.51 |
ENST00000374303.2
ENST00000533762.1 ENST00000529116.1 ENST00000474295.1 ENST00000488327.2 ENST00000472643.1 ENST00000526894.1 ENST00000524618.1 ENST00000374307.5 |
MTFR1L
|
mitochondrial fission regulator 1-like |
| chr2_+_48667898 | 0.50 |
ENST00000281394.4
ENST00000294952.8 |
PPP1R21
|
protein phosphatase 1, regulatory subunit 21 |
| chr3_+_111578027 | 0.50 |
ENST00000431670.2
ENST00000412622.1 |
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr19_-_46272106 | 0.50 |
ENST00000560168.1
|
SIX5
|
SIX homeobox 5 |
| chrX_-_114468605 | 0.48 |
ENST00000538422.1
ENST00000317135.8 |
LRCH2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr17_-_65241281 | 0.48 |
ENST00000358691.5
ENST00000580168.1 |
HELZ
|
helicase with zinc finger |
| chr12_+_109535373 | 0.48 |
ENST00000242576.2
|
UNG
|
uracil-DNA glycosylase |
| chr1_-_46598371 | 0.47 |
ENST00000372006.1
ENST00000425892.1 ENST00000420542.1 ENST00000354242.4 ENST00000340332.6 |
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr6_-_112575758 | 0.46 |
ENST00000431543.2
ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4
|
laminin, alpha 4 |
| chr9_+_79792269 | 0.46 |
ENST00000376634.4
ENST00000376636.3 ENST00000360280.3 |
VPS13A
|
vacuolar protein sorting 13 homolog A (S. cerevisiae) |
| chrX_-_133931164 | 0.46 |
ENST00000370790.1
ENST00000298090.6 |
FAM122B
|
family with sequence similarity 122B |
| chr3_-_129612394 | 0.46 |
ENST00000505616.1
ENST00000426664.2 |
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr21_-_34100244 | 0.45 |
ENST00000382491.3
ENST00000357345.3 ENST00000429236.1 |
SYNJ1
|
synaptojanin 1 |
| chr3_+_111578131 | 0.45 |
ENST00000498699.1
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr10_+_112404132 | 0.44 |
ENST00000369519.3
|
RBM20
|
RNA binding motif protein 20 |
| chr8_+_42396936 | 0.44 |
ENST00000416469.2
|
SMIM19
|
small integral membrane protein 19 |
| chr13_+_76210448 | 0.43 |
ENST00000377499.5
|
LMO7
|
LIM domain 7 |
| chr1_+_26146674 | 0.43 |
ENST00000525713.1
ENST00000374301.3 |
MTFR1L
|
mitochondrial fission regulator 1-like |
| chr10_-_69835099 | 0.43 |
ENST00000373700.4
|
HERC4
|
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
| chr11_-_18343669 | 0.42 |
ENST00000396253.3
ENST00000349215.3 ENST00000438420.2 |
HPS5
|
Hermansky-Pudlak syndrome 5 |
| chr8_-_27469196 | 0.42 |
ENST00000546343.1
ENST00000560566.1 |
CLU
|
clusterin |
| chr19_-_45909585 | 0.42 |
ENST00000593226.1
ENST00000418234.2 |
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like |
| chr16_+_2014993 | 0.42 |
ENST00000564014.1
|
SNHG9
|
small nucleolar RNA host gene 9 (non-protein coding) |
| chr1_+_180601139 | 0.42 |
ENST00000367590.4
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr2_-_105946491 | 0.42 |
ENST00000393359.2
|
TGFBRAP1
|
transforming growth factor, beta receptor associated protein 1 |
| chr10_+_38299546 | 0.41 |
ENST00000374618.3
ENST00000432900.2 ENST00000458705.2 ENST00000469037.2 |
ZNF33A
|
zinc finger protein 33A |
| chr2_+_220306745 | 0.41 |
ENST00000431523.1
ENST00000396698.1 ENST00000396695.2 |
SPEG
|
SPEG complex locus |
| chr6_-_31864977 | 0.40 |
ENST00000395728.3
ENST00000375528.4 |
EHMT2
|
euchromatic histone-lysine N-methyltransferase 2 |
| chr15_-_72490114 | 0.40 |
ENST00000309731.7
|
GRAMD2
|
GRAM domain containing 2 |
| chr3_+_111578640 | 0.40 |
ENST00000393925.3
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr3_-_123168551 | 0.40 |
ENST00000462833.1
|
ADCY5
|
adenylate cyclase 5 |
| chr13_-_76111945 | 0.40 |
ENST00000355801.4
ENST00000406936.3 |
COMMD6
|
COMM domain containing 6 |
| chr4_+_145567297 | 0.39 |
ENST00000434550.2
|
HHIP
|
hedgehog interacting protein |
| chr3_+_170075436 | 0.39 |
ENST00000476188.1
ENST00000259119.4 ENST00000426052.2 |
SKIL
|
SKI-like oncogene |
| chr16_-_10674528 | 0.39 |
ENST00000359543.3
|
EMP2
|
epithelial membrane protein 2 |
| chr2_-_201828356 | 0.38 |
ENST00000234296.2
|
ORC2
|
origin recognition complex, subunit 2 |
| chr11_+_67159416 | 0.38 |
ENST00000307980.2
ENST00000544620.1 |
RAD9A
|
RAD9 homolog A (S. pombe) |
| chrX_+_47444613 | 0.38 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr1_-_171711387 | 0.38 |
ENST00000236192.7
|
VAMP4
|
vesicle-associated membrane protein 4 |
| chr17_+_62075703 | 0.38 |
ENST00000577953.1
ENST00000582540.1 ENST00000579184.1 ENST00000425164.3 ENST00000412177.1 ENST00000539996.1 ENST00000583891.1 ENST00000580752.1 |
C17orf72
|
chromosome 17 open reading frame 72 |
| chr7_-_8302164 | 0.38 |
ENST00000447326.1
ENST00000406470.2 |
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr6_+_43149903 | 0.37 |
ENST00000252050.4
ENST00000354495.3 ENST00000372647.2 |
CUL9
|
cullin 9 |
| chr12_+_107168418 | 0.37 |
ENST00000392839.2
ENST00000548914.1 ENST00000355478.2 ENST00000552619.1 ENST00000549643.1 |
RIC8B
|
RIC8 guanine nucleotide exchange factor B |
| chr7_-_8301768 | 0.37 |
ENST00000265577.7
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr6_-_79787902 | 0.37 |
ENST00000275034.4
|
PHIP
|
pleckstrin homology domain interacting protein |
| chr5_+_111496631 | 0.36 |
ENST00000508590.1
|
EPB41L4A-AS1
|
EPB41L4A antisense RNA 1 |
| chr20_-_49253425 | 0.36 |
ENST00000045083.2
|
FAM65C
|
family with sequence similarity 65, member C |
| chr7_-_8301682 | 0.36 |
ENST00000396675.3
ENST00000430867.1 |
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr3_-_47517302 | 0.36 |
ENST00000441517.2
ENST00000545718.1 |
SCAP
|
SREBF chaperone |
| chr5_+_111496554 | 0.36 |
ENST00000442823.2
|
EPB41L4A-AS1
|
EPB41L4A antisense RNA 1 |
| chr5_+_121647386 | 0.36 |
ENST00000542191.1
ENST00000506272.1 ENST00000508681.1 ENST00000509154.2 |
SNCAIP
|
synuclein, alpha interacting protein |
| chr14_-_24804269 | 0.36 |
ENST00000310677.4
ENST00000554068.2 ENST00000559167.1 ENST00000561138.1 |
ADCY4
|
adenylate cyclase 4 |
| chr3_-_125094093 | 0.35 |
ENST00000484491.1
ENST00000492394.1 ENST00000471196.1 ENST00000468369.1 ENST00000544464.1 ENST00000485866.1 ENST00000360647.4 |
ZNF148
|
zinc finger protein 148 |
| chr8_+_42396712 | 0.35 |
ENST00000518574.1
ENST00000417410.2 ENST00000414154.2 |
SMIM19
|
small integral membrane protein 19 |
| chrX_-_119763835 | 0.35 |
ENST00000371313.2
ENST00000304661.5 |
C1GALT1C1
|
C1GALT1-specific chaperone 1 |
| chrX_+_130192216 | 0.35 |
ENST00000276211.5
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr2_-_43823093 | 0.35 |
ENST00000405006.4
|
THADA
|
thyroid adenoma associated |
| chr20_-_18038521 | 0.35 |
ENST00000278780.6
|
OVOL2
|
ovo-like zinc finger 2 |
| chr16_-_90085824 | 0.34 |
ENST00000002501.6
|
DBNDD1
|
dysbindin (dystrobrevin binding protein 1) domain containing 1 |
| chr10_-_123687497 | 0.34 |
ENST00000369040.3
ENST00000224652.6 ENST00000369043.3 |
ATE1
|
arginyltransferase 1 |
| chr4_+_150999418 | 0.34 |
ENST00000296550.7
|
DCLK2
|
doublecortin-like kinase 2 |
| chr12_+_27677085 | 0.34 |
ENST00000545334.1
ENST00000540114.1 ENST00000537927.1 ENST00000318304.8 ENST00000535047.1 ENST00000542629.1 ENST00000228425.6 |
PPFIBP1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
| chr17_-_42277203 | 0.34 |
ENST00000587097.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr1_+_95699704 | 0.33 |
ENST00000370202.4
|
RWDD3
|
RWD domain containing 3 |
| chr17_+_45608614 | 0.33 |
ENST00000544660.1
|
NPEPPS
|
aminopeptidase puromycin sensitive |
| chr4_-_151936416 | 0.33 |
ENST00000510413.1
ENST00000507224.1 |
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr20_-_46415341 | 0.32 |
ENST00000484875.1
ENST00000361612.4 |
SULF2
|
sulfatase 2 |
| chr20_-_46415297 | 0.32 |
ENST00000467815.1
ENST00000359930.4 |
SULF2
|
sulfatase 2 |
| chr2_+_32390925 | 0.32 |
ENST00000440718.1
ENST00000379343.2 ENST00000282587.5 ENST00000435660.1 ENST00000538303.1 ENST00000357055.3 ENST00000406369.1 |
SLC30A6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr21_-_34100341 | 0.31 |
ENST00000382499.2
ENST00000433931.2 |
SYNJ1
|
synaptojanin 1 |
| chr10_+_75532028 | 0.31 |
ENST00000372841.3
ENST00000394790.1 |
FUT11
|
fucosyltransferase 11 (alpha (1,3) fucosyltransferase) |
| chr15_+_91446157 | 0.31 |
ENST00000559717.1
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2 |
| chr7_-_8301869 | 0.31 |
ENST00000402384.3
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr2_+_204193149 | 0.31 |
ENST00000422511.2
|
ABI2
|
abl-interactor 2 |
| chr2_+_234545148 | 0.31 |
ENST00000373445.1
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr11_+_57508825 | 0.31 |
ENST00000534355.1
|
C11orf31
|
chromosome 11 open reading frame 31 |
| chr11_+_64794875 | 0.30 |
ENST00000377244.3
ENST00000534637.1 ENST00000524831.1 |
SNX15
|
sorting nexin 15 |
| chr1_-_87379785 | 0.30 |
ENST00000401030.3
ENST00000370554.1 |
SEP15
|
Homo sapiens 15 kDa selenoprotein (SEP15), transcript variant 2, mRNA. |
| chr16_+_15528332 | 0.29 |
ENST00000566490.1
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr12_+_49212514 | 0.29 |
ENST00000301050.2
ENST00000548279.1 ENST00000547230.1 |
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit |
| chr22_-_29663954 | 0.29 |
ENST00000216085.7
|
RHBDD3
|
rhomboid domain containing 3 |
| chr5_+_57878859 | 0.28 |
ENST00000282878.4
|
RAB3C
|
RAB3C, member RAS oncogene family |
| chr2_-_183903133 | 0.28 |
ENST00000361354.4
|
NCKAP1
|
NCK-associated protein 1 |
| chr17_-_73267214 | 0.28 |
ENST00000580717.1
ENST00000577542.1 ENST00000579612.1 ENST00000245551.5 |
MIF4GD
|
MIF4G domain containing |
| chr10_-_69834973 | 0.28 |
ENST00000395187.2
|
HERC4
|
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
| chr1_+_222817629 | 0.28 |
ENST00000340535.7
|
MIA3
|
melanoma inhibitory activity family, member 3 |
| chr5_+_121647764 | 0.28 |
ENST00000261368.8
ENST00000379533.2 ENST00000379536.2 ENST00000379538.3 |
SNCAIP
|
synuclein, alpha interacting protein |
| chr8_+_125486939 | 0.27 |
ENST00000303545.3
|
RNF139
|
ring finger protein 139 |
| chr10_-_17659357 | 0.27 |
ENST00000326961.6
ENST00000361271.3 |
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr9_+_96338647 | 0.27 |
ENST00000359246.4
|
PHF2
|
PHD finger protein 2 |
| chr5_+_159343688 | 0.26 |
ENST00000306675.3
|
ADRA1B
|
adrenoceptor alpha 1B |
| chr4_+_159593418 | 0.26 |
ENST00000507475.1
ENST00000307738.5 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr12_+_107168342 | 0.25 |
ENST00000392837.4
|
RIC8B
|
RIC8 guanine nucleotide exchange factor B |
| chr9_+_131451480 | 0.25 |
ENST00000322030.8
|
SET
|
SET nuclear oncogene |
| chr2_+_54198210 | 0.25 |
ENST00000607452.1
ENST00000422521.2 |
ACYP2
|
acylphosphatase 2, muscle type |
| chr2_+_204193129 | 0.25 |
ENST00000417864.1
|
ABI2
|
abl-interactor 2 |
| chr5_-_137368708 | 0.25 |
ENST00000033079.3
|
FAM13B
|
family with sequence similarity 13, member B |
| chr2_+_204192942 | 0.25 |
ENST00000295851.5
ENST00000261017.5 |
ABI2
|
abl-interactor 2 |
| chr17_-_73267281 | 0.25 |
ENST00000578305.1
ENST00000325102.8 |
MIF4GD
|
MIF4G domain containing |
| chr17_-_73267304 | 0.24 |
ENST00000579297.1
ENST00000580571.1 |
MIF4GD
|
MIF4G domain containing |
| chr5_-_42811986 | 0.24 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr22_-_38794490 | 0.24 |
ENST00000400206.2
|
CSNK1E
|
casein kinase 1, epsilon |
| chr1_-_94312706 | 0.24 |
ENST00000370244.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr1_-_46598284 | 0.24 |
ENST00000423209.1
ENST00000262741.5 |
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr7_+_142960505 | 0.23 |
ENST00000409500.3
ENST00000443571.2 ENST00000358406.5 ENST00000479303.1 |
GSTK1
|
glutathione S-transferase kappa 1 |
| chr5_+_175298674 | 0.23 |
ENST00000514150.1
|
CPLX2
|
complexin 2 |
| chr1_-_156390128 | 0.23 |
ENST00000368242.3
|
C1orf61
|
chromosome 1 open reading frame 61 |
| chr18_+_56338750 | 0.23 |
ENST00000345724.3
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1 |
| chr7_+_26438187 | 0.23 |
ENST00000439120.1
ENST00000430548.1 ENST00000421862.1 ENST00000449537.1 ENST00000420774.1 ENST00000418758.2 |
AC004540.5
|
AC004540.5 |
| chr20_+_3026591 | 0.23 |
ENST00000380325.3
|
MRPS26
|
mitochondrial ribosomal protein S26 |
| chr14_+_57857262 | 0.22 |
ENST00000555166.1
ENST00000556492.1 ENST00000554703.1 |
NAA30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chr2_+_234545092 | 0.22 |
ENST00000344644.5
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr17_-_28618948 | 0.22 |
ENST00000261714.6
|
BLMH
|
bleomycin hydrolase |
| chr17_-_36413133 | 0.22 |
ENST00000523089.1
ENST00000312412.4 ENST00000520237.1 |
RP11-1407O15.2
|
TBC1 domain family member 3 |
| chr11_+_111957497 | 0.21 |
ENST00000375549.3
ENST00000528182.1 ENST00000528048.1 ENST00000528021.1 ENST00000526592.1 ENST00000525291.1 |
SDHD
|
succinate dehydrogenase complex, subunit D, integral membrane protein |
| chr2_-_43823119 | 0.21 |
ENST00000403856.1
ENST00000404790.1 ENST00000405975.2 ENST00000415080.2 |
THADA
|
thyroid adenoma associated |
| chr14_+_96968707 | 0.21 |
ENST00000216277.8
ENST00000557320.1 ENST00000557471.1 |
PAPOLA
|
poly(A) polymerase alpha |
| chr1_+_52608046 | 0.21 |
ENST00000357206.2
ENST00000287727.3 |
ZFYVE9
|
zinc finger, FYVE domain containing 9 |
| chr19_+_56116771 | 0.21 |
ENST00000568956.1
|
ZNF865
|
zinc finger protein 865 |
| chr19_+_3572925 | 0.21 |
ENST00000333651.6
ENST00000417382.1 ENST00000453933.1 ENST00000262949.7 |
HMG20B
|
high mobility group 20B |
| chr2_-_38830160 | 0.20 |
ENST00000409636.1
ENST00000608859.1 ENST00000358367.4 |
HNRNPLL
|
heterogeneous nuclear ribonucleoprotein L-like |
| chr7_+_75677354 | 0.20 |
ENST00000461263.2
ENST00000315758.5 ENST00000443006.1 |
MDH2
|
malate dehydrogenase 2, NAD (mitochondrial) |
| chr11_-_117186946 | 0.20 |
ENST00000313005.6
ENST00000528053.1 |
BACE1
|
beta-site APP-cleaving enzyme 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR0B1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.1 | GO:0051622 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.5 | 3.4 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.5 | 3.9 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.5 | 1.8 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.4 | 1.1 | GO:0010645 | pulmonary valve formation(GO:0003193) regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) foramen ovale closure(GO:0035922) regulation of bundle of His cell action potential(GO:0098905) |
| 0.4 | 1.8 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.3 | 1.7 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.3 | 1.9 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.3 | 0.8 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.2 | 0.9 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.2 | 0.6 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.2 | 0.8 | GO:1904978 | regulation of endosome organization(GO:1904978) |
| 0.2 | 0.6 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.2 | 0.5 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.2 | 0.6 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.6 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.1 | 0.4 | GO:0070631 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.1 | 0.8 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 0.8 | GO:0010748 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.1 | 1.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 0.8 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 | 0.8 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 0.3 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.1 | 1.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.4 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.3 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 2.1 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 1.1 | GO:0003149 | membranous septum morphogenesis(GO:0003149) |
| 0.1 | 1.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 1.8 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 0.4 | GO:1902847 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 0.3 | GO:1901091 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.1 | 1.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.7 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.1 | 0.2 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.3 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.1 | 0.2 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.1 | 0.2 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.1 | 0.4 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.1 | 0.3 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.1 | 0.3 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.1 | 0.2 | GO:1903823 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.1 | 0.7 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 0.2 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.1 | 0.2 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.1 | 0.9 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 0.4 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.7 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 1.3 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.2 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.0 | 0.2 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.0 | 0.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.8 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.4 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.2 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.0 | 0.3 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 1.0 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.4 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 1.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.4 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.4 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.3 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.3 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.8 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.6 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 1.7 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.3 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:1903567 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.5 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.3 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.3 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 0.7 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.1 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.7 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.0 | 0.5 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:1900114 | pathogenesis(GO:0009405) positive regulation of histone H3-K9 trimethylation(GO:1900114) |
| 0.0 | 0.1 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.0 | 2.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 1.3 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.0 | 0.2 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.8 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.9 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.7 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.2 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.2 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.5 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.0 | 0.1 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.2 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.4 | GO:0036344 | platelet morphogenesis(GO:0036344) |
| 0.0 | 0.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.3 | GO:0036065 | fucosylation(GO:0036065) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 3.4 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.4 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 1.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.4 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.1 | 0.4 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.1 | 2.0 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.9 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 0.8 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 3.9 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 3.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.4 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 0.3 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 1.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.2 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 0.2 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 1.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.4 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 1.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.9 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.3 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.4 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.8 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 1.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.2 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 2.6 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 1.0 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.8 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 1.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 1.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.1 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 2.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.6 | 3.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.6 | 3.9 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.3 | 2.5 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.3 | 1.1 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.2 | 0.7 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 0.9 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 1.8 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.2 | 0.9 | GO:0004057 | arginyltransferase activity(GO:0004057) |
| 0.2 | 0.8 | GO:0043398 | HLH domain binding(GO:0043398) calcium-induced calcium release activity(GO:0048763) |
| 0.2 | 0.8 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 1.0 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.9 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.4 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 1.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.6 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.3 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.2 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.1 | 0.2 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.2 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.1 | 0.4 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.4 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.5 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.1 | 0.3 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 0.2 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 1.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.2 | GO:0043813 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.0 | 0.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 1.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.5 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.3 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 2.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.2 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 1.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.8 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.2 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 1.0 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 1.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.4 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 1.0 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 1.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.2 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 3.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 2.0 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.7 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.8 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.8 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.9 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 0.8 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.8 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.8 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 3.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.4 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 1.0 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.4 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 1.4 | REACTOME DNA REPAIR | Genes involved in DNA Repair |