Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
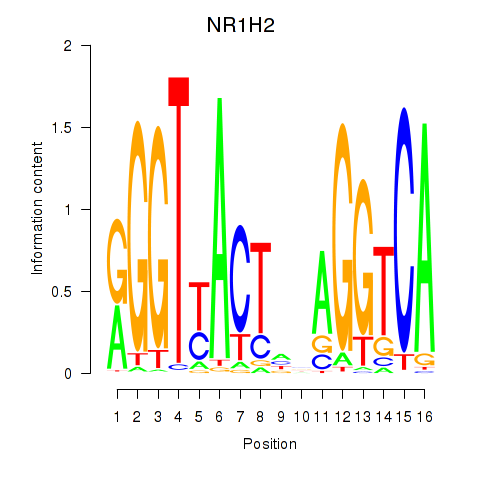
Results for NR1H2
Z-value: 0.47
Transcription factors associated with NR1H2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1H2
|
ENSG00000131408.9 | nuclear receptor subfamily 1 group H member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR1H2 | hg19_v2_chr19_+_50832943_50832951 | 0.17 | 4.2e-01 | Click! |
Activity profile of NR1H2 motif
Sorted Z-values of NR1H2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_43619796 | 1.78 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr1_+_169075554 | 1.38 |
ENST00000367815.4
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr17_+_77019030 | 1.28 |
ENST00000580454.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr17_+_77018896 | 1.25 |
ENST00000578229.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chrX_+_1455478 | 1.18 |
ENST00000331035.4
|
IL3RA
|
interleukin 3 receptor, alpha (low affinity) |
| chr4_-_681114 | 1.09 |
ENST00000503156.1
|
MFSD7
|
major facilitator superfamily domain containing 7 |
| chr7_-_22259845 | 1.05 |
ENST00000420196.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr2_+_223725652 | 1.04 |
ENST00000357430.3
ENST00000392066.3 |
ACSL3
|
acyl-CoA synthetase long-chain family member 3 |
| chr5_+_35856951 | 0.90 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr20_+_31805131 | 0.86 |
ENST00000375454.3
ENST00000375452.3 |
BPIFA3
|
BPI fold containing family A, member 3 |
| chr19_-_49658387 | 0.70 |
ENST00000595625.1
|
HRC
|
histidine rich calcium binding protein |
| chr9_-_139293249 | 0.66 |
ENST00000298532.2
|
SNAPC4
|
small nuclear RNA activating complex, polypeptide 4, 190kDa |
| chr1_-_237167718 | 0.66 |
ENST00000464121.2
|
MT1HL1
|
metallothionein 1H-like 1 |
| chr5_-_138862326 | 0.63 |
ENST00000330794.4
|
TMEM173
|
transmembrane protein 173 |
| chr18_+_21693306 | 0.60 |
ENST00000540918.2
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr2_+_73441350 | 0.50 |
ENST00000389501.4
|
SMYD5
|
SMYD family member 5 |
| chr19_+_33071974 | 0.48 |
ENST00000590247.2
ENST00000419343.3 ENST00000592786.1 ENST00000379316.3 |
PDCD5
|
programmed cell death 5 |
| chr16_+_4674814 | 0.47 |
ENST00000415496.1
ENST00000587747.1 ENST00000399577.5 ENST00000588994.1 ENST00000586183.1 |
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr7_-_76247617 | 0.46 |
ENST00000441393.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr11_-_82611448 | 0.43 |
ENST00000393399.2
ENST00000313010.3 |
PRCP
|
prolylcarboxypeptidase (angiotensinase C) |
| chr1_+_23037323 | 0.42 |
ENST00000544305.1
ENST00000374630.3 ENST00000400191.3 ENST00000374632.3 |
EPHB2
|
EPH receptor B2 |
| chr17_-_49198216 | 0.41 |
ENST00000262013.7
ENST00000357122.4 |
SPAG9
|
sperm associated antigen 9 |
| chr16_+_4674787 | 0.41 |
ENST00000262370.7
|
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr1_-_161207986 | 0.40 |
ENST00000506209.1
ENST00000367980.2 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr15_+_75498739 | 0.39 |
ENST00000565074.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr8_-_48651648 | 0.39 |
ENST00000408965.3
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr17_-_8151353 | 0.39 |
ENST00000315684.8
|
CTC1
|
CTS telomere maintenance complex component 1 |
| chr22_-_24096630 | 0.38 |
ENST00000248948.3
|
VPREB3
|
pre-B lymphocyte 3 |
| chr1_-_161207953 | 0.38 |
ENST00000367982.4
|
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr1_-_161208013 | 0.38 |
ENST00000515452.1
ENST00000367983.4 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr20_-_5485166 | 0.37 |
ENST00000589201.1
ENST00000379053.4 |
LINC00654
|
long intergenic non-protein coding RNA 654 |
| chr7_+_153584166 | 0.36 |
ENST00000404039.1
|
DPP6
|
dipeptidyl-peptidase 6 |
| chr17_+_32683456 | 0.33 |
ENST00000225844.2
|
CCL13
|
chemokine (C-C motif) ligand 13 |
| chr12_-_322504 | 0.31 |
ENST00000424061.2
|
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr2_+_168043793 | 0.31 |
ENST00000409273.1
ENST00000409605.1 |
XIRP2
|
xin actin-binding repeat containing 2 |
| chr17_-_17480779 | 0.31 |
ENST00000395782.1
|
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chrX_+_23801280 | 0.30 |
ENST00000379251.3
ENST00000379253.3 ENST00000379254.1 ENST00000379270.4 |
SAT1
|
spermidine/spermine N1-acetyltransferase 1 |
| chr2_+_27255806 | 0.30 |
ENST00000238788.9
ENST00000404032.3 |
TMEM214
|
transmembrane protein 214 |
| chr12_-_322821 | 0.30 |
ENST00000359674.4
|
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr9_-_130829588 | 0.29 |
ENST00000373078.4
|
NAIF1
|
nuclear apoptosis inducing factor 1 |
| chr3_-_127309879 | 0.28 |
ENST00000489960.1
ENST00000490290.1 |
TPRA1
|
transmembrane protein, adipocyte asscociated 1 |
| chr10_-_70287172 | 0.27 |
ENST00000539557.1
|
SLC25A16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr22_-_24096562 | 0.27 |
ENST00000398465.3
|
VPREB3
|
pre-B lymphocyte 3 |
| chr3_-_127309550 | 0.27 |
ENST00000296210.7
ENST00000355552.3 |
TPRA1
|
transmembrane protein, adipocyte asscociated 1 |
| chr15_-_89010607 | 0.26 |
ENST00000312475.4
|
MRPL46
|
mitochondrial ribosomal protein L46 |
| chr11_-_62420757 | 0.26 |
ENST00000330574.2
|
INTS5
|
integrator complex subunit 5 |
| chr1_+_87595542 | 0.25 |
ENST00000461990.1
|
RP5-1052I5.1
|
long intergenic non-protein coding RNA 1140 |
| chr20_+_33292068 | 0.25 |
ENST00000374810.3
ENST00000374809.2 ENST00000451665.1 |
TP53INP2
|
tumor protein p53 inducible nuclear protein 2 |
| chr17_-_19648916 | 0.25 |
ENST00000444455.1
ENST00000439102.2 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr16_+_56969284 | 0.25 |
ENST00000568358.1
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr19_+_34287174 | 0.25 |
ENST00000587559.1
ENST00000588637.1 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr16_-_4465886 | 0.25 |
ENST00000539968.1
|
CORO7
|
coronin 7 |
| chr3_-_195310802 | 0.25 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr3_+_9691117 | 0.23 |
ENST00000353332.5
ENST00000420925.1 ENST00000296003.4 ENST00000351233.5 |
MTMR14
|
myotubularin related protein 14 |
| chr12_-_9913489 | 0.23 |
ENST00000228434.3
ENST00000536709.1 |
CD69
|
CD69 molecule |
| chr1_-_9714644 | 0.23 |
ENST00000377320.3
|
C1orf200
|
chromosome 1 open reading frame 200 |
| chr15_+_81589254 | 0.23 |
ENST00000394652.2
|
IL16
|
interleukin 16 |
| chr7_+_142031986 | 0.22 |
ENST00000547918.2
|
TRBV7-1
|
T cell receptor beta variable 7-1 (non-functional) |
| chr4_-_2965052 | 0.22 |
ENST00000398071.4
ENST00000502735.1 ENST00000314262.6 ENST00000416614.2 |
NOP14
|
NOP14 nucleolar protein |
| chr16_+_30710462 | 0.21 |
ENST00000262518.4
ENST00000395059.2 ENST00000344771.4 |
SRCAP
|
Snf2-related CREBBP activator protein |
| chr11_+_1074870 | 0.21 |
ENST00000441003.2
ENST00000359061.5 |
MUC2
|
mucin 2, oligomeric mucus/gel-forming |
| chr1_+_10459111 | 0.21 |
ENST00000541529.1
ENST00000270776.8 ENST00000483936.1 ENST00000538557.1 |
PGD
|
phosphogluconate dehydrogenase |
| chr19_-_46285646 | 0.21 |
ENST00000458663.2
|
DMPK
|
dystrophia myotonica-protein kinase |
| chr8_+_132916318 | 0.21 |
ENST00000254624.5
ENST00000522709.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr1_+_156338993 | 0.20 |
ENST00000368249.1
ENST00000368246.2 ENST00000537040.1 ENST00000400992.2 ENST00000255013.3 ENST00000451864.2 |
RHBG
|
Rh family, B glycoprotein (gene/pseudogene) |
| chr4_+_57371509 | 0.20 |
ENST00000360096.2
|
ARL9
|
ADP-ribosylation factor-like 9 |
| chr19_+_41092680 | 0.19 |
ENST00000594298.1
ENST00000597396.1 |
SHKBP1
|
SH3KBP1 binding protein 1 |
| chr20_+_43343886 | 0.19 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr17_-_42580738 | 0.19 |
ENST00000585614.1
ENST00000591680.1 ENST00000434000.1 ENST00000588554.1 ENST00000592154.1 |
GPATCH8
|
G patch domain containing 8 |
| chr11_+_77774897 | 0.19 |
ENST00000281030.2
|
THRSP
|
thyroid hormone responsive |
| chr15_-_102285007 | 0.19 |
ENST00000560292.2
|
RP11-89K11.1
|
Uncharacterized protein |
| chr12_-_56694142 | 0.18 |
ENST00000550655.1
ENST00000548567.1 ENST00000551430.2 ENST00000351328.3 |
CS
|
citrate synthase |
| chr12_+_56511943 | 0.18 |
ENST00000257940.2
ENST00000552345.1 ENST00000551880.1 ENST00000546903.1 ENST00000551790.1 |
ZC3H10
ESYT1
|
zinc finger CCCH-type containing 10 extended synaptotagmin-like protein 1 |
| chr2_-_165697920 | 0.18 |
ENST00000342193.4
ENST00000375458.2 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr6_+_30687978 | 0.18 |
ENST00000327892.8
ENST00000435534.1 |
TUBB
|
tubulin, beta class I |
| chr1_-_11007927 | 0.18 |
ENST00000468348.1
|
C1orf127
|
chromosome 1 open reading frame 127 |
| chr11_-_62521614 | 0.18 |
ENST00000527994.1
ENST00000394807.3 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chr1_+_10290822 | 0.18 |
ENST00000377083.1
|
KIF1B
|
kinesin family member 1B |
| chr17_-_8198636 | 0.18 |
ENST00000577745.1
ENST00000579192.1 ENST00000396278.1 |
SLC25A35
|
solute carrier family 25, member 35 |
| chr21_-_46340770 | 0.18 |
ENST00000397854.3
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr11_+_64692143 | 0.18 |
ENST00000164133.2
ENST00000532850.1 |
PPP2R5B
|
protein phosphatase 2, regulatory subunit B', beta |
| chr19_+_52264104 | 0.18 |
ENST00000340023.6
|
FPR2
|
formyl peptide receptor 2 |
| chr22_+_18260077 | 0.18 |
ENST00000600723.1
|
LINC00528
|
long intergenic non-protein coding RNA 528 |
| chr20_+_43343517 | 0.17 |
ENST00000372865.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr7_-_56160625 | 0.17 |
ENST00000446428.1
ENST00000432123.1 ENST00000452681.2 ENST00000537360.1 |
PHKG1
|
phosphorylase kinase, gamma 1 (muscle) |
| chr20_+_5987890 | 0.17 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr5_+_43120985 | 0.17 |
ENST00000515326.1
|
ZNF131
|
zinc finger protein 131 |
| chr1_+_100818156 | 0.16 |
ENST00000336454.3
|
CDC14A
|
cell division cycle 14A |
| chr16_+_577697 | 0.16 |
ENST00000562370.1
ENST00000568988.1 ENST00000219611.2 |
CAPN15
|
calpain 15 |
| chr22_+_30792846 | 0.16 |
ENST00000312932.9
ENST00000428195.1 |
SEC14L2
|
SEC14-like 2 (S. cerevisiae) |
| chr3_-_49893958 | 0.16 |
ENST00000482243.1
ENST00000331456.2 ENST00000469027.1 |
TRAIP
|
TRAF interacting protein |
| chr3_+_39371255 | 0.16 |
ENST00000414803.1
ENST00000545843.1 |
CCR8
|
chemokine (C-C motif) receptor 8 |
| chr16_+_29467780 | 0.16 |
ENST00000395400.3
|
SULT1A4
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4 |
| chrX_+_30248553 | 0.15 |
ENST00000361644.2
|
MAGEB3
|
melanoma antigen family B, 3 |
| chr3_+_39371191 | 0.15 |
ENST00000326306.4
|
CCR8
|
chemokine (C-C motif) receptor 8 |
| chr11_-_35440796 | 0.14 |
ENST00000278379.3
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr19_+_3224700 | 0.14 |
ENST00000292672.2
ENST00000541430.2 |
CELF5
|
CUGBP, Elav-like family member 5 |
| chr1_-_67142710 | 0.14 |
ENST00000502413.2
|
AL139147.1
|
Uncharacterized protein |
| chr2_+_145425573 | 0.14 |
ENST00000600064.1
ENST00000597670.1 ENST00000414256.1 ENST00000599187.1 ENST00000451774.1 ENST00000599072.1 ENST00000596589.1 ENST00000597893.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr7_-_56160666 | 0.14 |
ENST00000297373.2
|
PHKG1
|
phosphorylase kinase, gamma 1 (muscle) |
| chr14_-_73925225 | 0.13 |
ENST00000356296.4
ENST00000355058.3 ENST00000359560.3 ENST00000557597.1 ENST00000554394.1 ENST00000555238.1 ENST00000535282.1 ENST00000555987.1 ENST00000555394.1 ENST00000554546.1 |
NUMB
|
numb homolog (Drosophila) |
| chr4_-_80247162 | 0.13 |
ENST00000286794.4
|
NAA11
|
N(alpha)-acetyltransferase 11, NatA catalytic subunit |
| chr4_-_83483395 | 0.13 |
ENST00000515780.2
|
TMEM150C
|
transmembrane protein 150C |
| chr5_-_58652788 | 0.13 |
ENST00000405755.2
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr11_+_32851487 | 0.13 |
ENST00000257836.3
|
PRRG4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr4_+_183164574 | 0.13 |
ENST00000511685.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr20_+_35918035 | 0.13 |
ENST00000373606.3
ENST00000397156.3 ENST00000397150.1 ENST00000397152.3 |
MANBAL
|
mannosidase, beta A, lysosomal-like |
| chr1_+_100818009 | 0.12 |
ENST00000370125.2
ENST00000361544.6 ENST00000370124.3 |
CDC14A
|
cell division cycle 14A |
| chr1_+_87595497 | 0.12 |
ENST00000471417.1
|
RP5-1052I5.1
|
long intergenic non-protein coding RNA 1140 |
| chr19_-_7968427 | 0.12 |
ENST00000539278.1
|
AC010336.1
|
Uncharacterized protein |
| chr17_-_26695013 | 0.12 |
ENST00000555059.2
|
CTB-96E2.2
|
Homeobox protein SEBOX |
| chr22_-_26875345 | 0.12 |
ENST00000398141.1
|
HPS4
|
Hermansky-Pudlak syndrome 4 |
| chr3_+_46283863 | 0.11 |
ENST00000545097.1
ENST00000541018.1 |
CCR3
|
chemokine (C-C motif) receptor 3 |
| chr1_+_169337172 | 0.11 |
ENST00000367807.3
ENST00000367808.3 ENST00000329281.2 ENST00000420531.1 |
BLZF1
|
basic leucine zipper nuclear factor 1 |
| chr4_-_142134031 | 0.11 |
ENST00000420921.2
|
RNF150
|
ring finger protein 150 |
| chr17_-_26694979 | 0.11 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr11_-_35441524 | 0.11 |
ENST00000395750.1
ENST00000449068.1 |
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr7_+_45067265 | 0.11 |
ENST00000474617.1
|
CCM2
|
cerebral cavernous malformation 2 |
| chr8_-_61193947 | 0.11 |
ENST00000317995.4
|
CA8
|
carbonic anhydrase VIII |
| chr4_-_39034542 | 0.11 |
ENST00000344606.6
|
TMEM156
|
transmembrane protein 156 |
| chr1_+_36335351 | 0.11 |
ENST00000373206.1
|
AGO1
|
argonaute RISC catalytic component 1 |
| chr2_+_207630081 | 0.10 |
ENST00000236980.6
ENST00000418289.1 ENST00000402774.3 ENST00000403094.3 |
FASTKD2
|
FAST kinase domains 2 |
| chr16_+_88704978 | 0.10 |
ENST00000244241.4
|
IL17C
|
interleukin 17C |
| chr9_+_95736758 | 0.10 |
ENST00000337352.6
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr15_-_83837983 | 0.10 |
ENST00000562702.1
|
HDGFRP3
|
Hepatoma-derived growth factor-related protein 3 |
| chr10_+_1095416 | 0.09 |
ENST00000358220.1
|
WDR37
|
WD repeat domain 37 |
| chr1_+_87458692 | 0.09 |
ENST00000370548.2
ENST00000356813.4 |
RP5-1052I5.2
HS2ST1
|
Heparan sulfate 2-O-sulfotransferase 1 heparan sulfate 2-O-sulfotransferase 1 |
| chr6_-_31763721 | 0.09 |
ENST00000375663.3
|
VARS
|
valyl-tRNA synthetase |
| chr9_-_86955657 | 0.09 |
ENST00000537648.1
|
SLC28A3
|
solute carrier family 28 (concentrative nucleoside transporter), member 3 |
| chr19_-_22193731 | 0.09 |
ENST00000601773.1
ENST00000397126.4 ENST00000601993.1 ENST00000599916.1 |
ZNF208
|
zinc finger protein 208 |
| chr9_+_95726243 | 0.09 |
ENST00000416701.2
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr6_-_34639733 | 0.09 |
ENST00000374021.1
|
C6orf106
|
chromosome 6 open reading frame 106 |
| chr14_-_59950724 | 0.09 |
ENST00000481608.1
|
L3HYPDH
|
L-3-hydroxyproline dehydratase (trans-) |
| chr21_-_37432540 | 0.09 |
ENST00000443703.1
ENST00000399207.1 ENST00000399215.1 ENST00000442559.1 ENST00000399205.1 ENST00000429161.1 ENST00000424303.1 ENST00000399208.2 |
SETD4
|
SET domain containing 4 |
| chr10_+_126490354 | 0.09 |
ENST00000298492.5
|
FAM175B
|
family with sequence similarity 175, member B |
| chr2_+_101179152 | 0.09 |
ENST00000264254.6
|
PDCL3
|
phosducin-like 3 |
| chr8_+_145218673 | 0.08 |
ENST00000326134.5
|
MROH1
|
maestro heat-like repeat family member 1 |
| chr17_+_7184986 | 0.07 |
ENST00000317370.8
ENST00000571308.1 |
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr17_-_76975925 | 0.07 |
ENST00000591274.1
ENST00000589906.1 ENST00000591778.1 ENST00000589775.2 ENST00000585407.1 ENST00000262776.3 |
LGALS3BP
|
lectin, galactoside-binding, soluble, 3 binding protein |
| chr7_-_73153122 | 0.07 |
ENST00000458339.1
|
ABHD11
|
abhydrolase domain containing 11 |
| chr19_-_46296011 | 0.07 |
ENST00000377735.3
ENST00000270223.6 |
DMWD
|
dystrophia myotonica, WD repeat containing |
| chr8_-_145652336 | 0.07 |
ENST00000529182.1
ENST00000526054.1 |
VPS28
|
vacuolar protein sorting 28 homolog (S. cerevisiae) |
| chr2_-_128615681 | 0.07 |
ENST00000409955.1
ENST00000272645.4 |
POLR2D
|
polymerase (RNA) II (DNA directed) polypeptide D |
| chrX_-_46759138 | 0.07 |
ENST00000377879.3
|
CXorf31
|
chromosome X open reading frame 31 |
| chr9_+_34179003 | 0.07 |
ENST00000545103.1
ENST00000543944.1 ENST00000536252.1 ENST00000540348.1 ENST00000297661.4 ENST00000379186.4 |
UBAP1
|
ubiquitin associated protein 1 |
| chr2_-_32390801 | 0.06 |
ENST00000608489.1
|
RP11-563N4.1
|
RP11-563N4.1 |
| chr5_-_133968529 | 0.06 |
ENST00000402673.2
|
SAR1B
|
SAR1 homolog B (S. cerevisiae) |
| chr11_-_35441597 | 0.06 |
ENST00000395753.1
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr15_+_89010923 | 0.06 |
ENST00000353598.6
|
MRPS11
|
mitochondrial ribosomal protein S11 |
| chr11_+_61159832 | 0.06 |
ENST00000334888.5
ENST00000398979.3 |
TMEM216
|
transmembrane protein 216 |
| chr4_-_71705027 | 0.06 |
ENST00000545193.1
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr14_-_102976135 | 0.06 |
ENST00000560748.1
|
ANKRD9
|
ankyrin repeat domain 9 |
| chr14_+_23790655 | 0.05 |
ENST00000397276.2
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr1_-_13115578 | 0.05 |
ENST00000414205.2
|
PRAMEF6
|
PRAME family member 6 |
| chr22_+_19118321 | 0.05 |
ENST00000399635.2
|
TSSK2
|
testis-specific serine kinase 2 |
| chr2_+_228029281 | 0.05 |
ENST00000396578.3
|
COL4A3
|
collagen, type IV, alpha 3 (Goodpasture antigen) |
| chr19_+_33622996 | 0.05 |
ENST00000592765.1
ENST00000361680.2 ENST00000355868.3 |
WDR88
|
WD repeat domain 88 |
| chr3_+_139062838 | 0.05 |
ENST00000310776.4
ENST00000465056.1 ENST00000465373.1 |
MRPS22
|
mitochondrial ribosomal protein S22 |
| chr4_-_71705060 | 0.05 |
ENST00000514161.1
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr10_-_28623368 | 0.05 |
ENST00000441595.2
|
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr2_+_145425534 | 0.05 |
ENST00000432608.1
ENST00000597655.1 ENST00000598659.1 ENST00000600679.1 ENST00000601277.1 ENST00000451027.1 ENST00000445791.1 ENST00000596540.1 ENST00000596230.1 ENST00000594471.1 ENST00000598248.1 ENST00000597469.1 ENST00000431734.1 ENST00000595686.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr17_-_15370915 | 0.04 |
ENST00000312177.6
|
CDRT4
|
CMT1A duplicated region transcript 4 |
| chr19_+_58545434 | 0.04 |
ENST00000282326.1
ENST00000601162.1 |
ZSCAN1
|
zinc finger and SCAN domain containing 1 |
| chr5_+_76145920 | 0.04 |
ENST00000317593.4
|
S100Z
|
S100 calcium binding protein Z |
| chr17_+_60457914 | 0.04 |
ENST00000305286.3
ENST00000520404.1 ENST00000518576.1 |
EFCAB3
|
EF-hand calcium binding domain 3 |
| chr22_-_36031181 | 0.04 |
ENST00000594060.1
|
AL049747.1
|
AL049747.1 |
| chr20_+_30028322 | 0.04 |
ENST00000376309.3
|
DEFB123
|
defensin, beta 123 |
| chr16_-_30023615 | 0.04 |
ENST00000564979.1
ENST00000563378.1 |
DOC2A
|
double C2-like domains, alpha |
| chr17_-_27054952 | 0.04 |
ENST00000580518.1
|
TLCD1
|
TLC domain containing 1 |
| chr4_+_5527117 | 0.04 |
ENST00000505296.1
|
C4orf6
|
chromosome 4 open reading frame 6 |
| chr19_+_15904761 | 0.04 |
ENST00000308940.8
|
OR10H5
|
olfactory receptor, family 10, subfamily H, member 5 |
| chr12_+_120972606 | 0.04 |
ENST00000413266.2
|
RNF10
|
ring finger protein 10 |
| chr19_+_58361185 | 0.04 |
ENST00000339656.5
ENST00000423137.1 |
ZNF587
|
zinc finger protein 587 |
| chr13_+_49684445 | 0.03 |
ENST00000398316.3
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr7_-_73153161 | 0.03 |
ENST00000395147.4
|
ABHD11
|
abhydrolase domain containing 11 |
| chr6_-_31763408 | 0.03 |
ENST00000444930.2
|
VARS
|
valyl-tRNA synthetase |
| chr9_-_86955598 | 0.03 |
ENST00000376238.4
|
SLC28A3
|
solute carrier family 28 (concentrative nucleoside transporter), member 3 |
| chr20_+_44330651 | 0.03 |
ENST00000305479.2
|
WFDC13
|
WAP four-disulfide core domain 13 |
| chr10_-_127511658 | 0.03 |
ENST00000368774.1
ENST00000368778.3 |
UROS
|
uroporphyrinogen III synthase |
| chr7_-_92219698 | 0.03 |
ENST00000438306.1
ENST00000445716.1 |
FAM133B
|
family with sequence similarity 133, member B |
| chr2_+_71357744 | 0.03 |
ENST00000498451.2
|
MPHOSPH10
|
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr22_+_36113919 | 0.03 |
ENST00000249044.2
|
APOL5
|
apolipoprotein L, 5 |
| chr2_+_113735575 | 0.02 |
ENST00000376489.2
ENST00000259205.4 |
IL36G
|
interleukin 36, gamma |
| chr16_-_68034470 | 0.02 |
ENST00000412757.2
|
DPEP2
|
dipeptidase 2 |
| chr15_+_64386261 | 0.02 |
ENST00000560829.1
|
SNX1
|
sorting nexin 1 |
| chr17_-_79604075 | 0.02 |
ENST00000374747.5
ENST00000539314.1 ENST00000331134.6 |
NPLOC4
|
nuclear protein localization 4 homolog (S. cerevisiae) |
| chr21_-_37432832 | 0.02 |
ENST00000332131.4
|
SETD4
|
SET domain containing 4 |
| chr15_+_91473403 | 0.01 |
ENST00000394275.2
|
UNC45A
|
unc-45 homolog A (C. elegans) |
| chr5_+_172410757 | 0.01 |
ENST00000519374.1
ENST00000519911.1 ENST00000265093.4 ENST00000517669.1 |
ATP6V0E1
|
ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 |
| chr14_-_77787198 | 0.01 |
ENST00000261534.4
|
POMT2
|
protein-O-mannosyltransferase 2 |
| chr14_-_94970559 | 0.01 |
ENST00000556881.1
|
SERPINA12
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 |
| chrX_-_9001351 | 0.01 |
ENST00000362066.3
|
FAM9B
|
family with sequence similarity 9, member B |
| chr5_+_76145826 | 0.01 |
ENST00000513010.1
|
S100Z
|
S100 calcium binding protein Z |
| chr1_+_13736907 | 0.01 |
ENST00000316412.5
|
PRAMEF20
|
PRAME family member 20 |
| chr4_-_39033963 | 0.01 |
ENST00000381938.3
|
TMEM156
|
transmembrane protein 156 |
| chr11_-_506316 | 0.01 |
ENST00000532055.1
ENST00000531540.1 |
RNH1
|
ribonuclease/angiogenin inhibitor 1 |
| chr13_+_23993097 | 0.00 |
ENST00000443092.1
|
SACS-AS1
|
SACS antisense RNA 1 |
| chr13_-_33112956 | 0.00 |
ENST00000505213.1
|
N4BP2L2
|
NEDD4 binding protein 2-like 2 |
| chr17_+_78152274 | 0.00 |
ENST00000344227.2
ENST00000570421.1 |
CARD14
|
caspase recruitment domain family, member 14 |
| chr1_-_153950098 | 0.00 |
ENST00000356648.1
|
JTB
|
jumping translocation breakpoint |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1H2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.3 | 1.0 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.3 | 2.5 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.3 | 1.4 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 0.2 | 0.7 | GO:1902081 | positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.2 | 1.2 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.2 | 0.7 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 0.5 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 0.4 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.1 | 0.2 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.5 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.1 | 0.3 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.1 | 0.9 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 0.9 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.2 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.1 | 0.2 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 0.2 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 0.4 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 0.4 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.4 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.0 | 0.2 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.0 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.0 | 0.3 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.0 | 0.1 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.6 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.6 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.4 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.1 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.0 | 0.2 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.1 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.2 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.3 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 1.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.7 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 2.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.1 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.3 | 0.9 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.2 | 1.1 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.2 | 0.5 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 0.3 | GO:0004608 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.1 | 0.6 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.3 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 1.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.2 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.1 | 1.0 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.2 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.1 | 0.7 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 0.3 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.5 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.1 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0047661 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.0 | 0.8 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.4 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.4 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 2.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.0 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 2.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID IL3 PATHWAY | IL3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.9 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.7 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |