Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
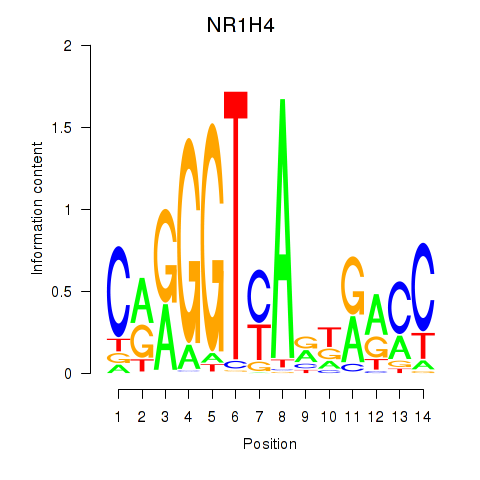
Results for NR1H4
Z-value: 0.48
Transcription factors associated with NR1H4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1H4
|
ENSG00000012504.9 | nuclear receptor subfamily 1 group H member 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR1H4 | hg19_v2_chr12_+_100867486_100867652 | 0.41 | 4.1e-02 | Click! |
Activity profile of NR1H4 motif
Sorted Z-values of NR1H4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_53809473 | 0.62 |
ENST00000575734.1
|
TMEM100
|
transmembrane protein 100 |
| chr8_+_9046503 | 0.54 |
ENST00000512942.2
|
RP11-10A14.5
|
RP11-10A14.5 |
| chr12_-_57914275 | 0.49 |
ENST00000547303.1
ENST00000552740.1 ENST00000547526.1 ENST00000551116.1 ENST00000346473.3 |
DDIT3
|
DNA-damage-inducible transcript 3 |
| chr17_+_59529743 | 0.47 |
ENST00000589003.1
ENST00000393853.4 |
TBX4
|
T-box 4 |
| chr11_+_7534999 | 0.44 |
ENST00000528947.1
ENST00000299492.4 |
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr20_-_45981138 | 0.44 |
ENST00000446994.2
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr1_+_162351503 | 0.44 |
ENST00000458626.2
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr10_-_95360983 | 0.39 |
ENST00000371464.3
|
RBP4
|
retinol binding protein 4, plasma |
| chr9_-_34372830 | 0.38 |
ENST00000379142.3
|
KIAA1161
|
KIAA1161 |
| chr9_-_137809718 | 0.35 |
ENST00000371806.3
|
FCN1
|
ficolin (collagen/fibrinogen domain containing) 1 |
| chr1_+_14026722 | 0.35 |
ENST00000376048.5
|
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr1_+_36789335 | 0.35 |
ENST00000373137.2
|
RP11-268J15.5
|
RP11-268J15.5 |
| chr2_-_106015527 | 0.35 |
ENST00000344213.4
ENST00000358129.4 |
FHL2
|
four and a half LIM domains 2 |
| chr12_+_109273806 | 0.32 |
ENST00000228476.3
ENST00000547768.1 |
DAO
|
D-amino-acid oxidase |
| chr3_+_118866222 | 0.32 |
ENST00000490594.1
|
RP11-484M3.5
|
Uncharacterized protein |
| chr17_-_74023291 | 0.31 |
ENST00000586740.1
|
EVPL
|
envoplakin |
| chr1_+_26560676 | 0.31 |
ENST00000451429.2
ENST00000252992.4 |
CEP85
|
centrosomal protein 85kDa |
| chr12_-_7281469 | 0.30 |
ENST00000542370.1
ENST00000266560.3 |
RBP5
|
retinol binding protein 5, cellular |
| chr14_-_21492251 | 0.30 |
ENST00000554398.1
|
NDRG2
|
NDRG family member 2 |
| chr14_-_21492113 | 0.29 |
ENST00000554094.1
|
NDRG2
|
NDRG family member 2 |
| chr17_+_4853442 | 0.29 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr2_+_233877324 | 0.28 |
ENST00000409905.1
|
AC106876.2
|
Uncharacterized protein |
| chr3_-_160117301 | 0.27 |
ENST00000326448.7
ENST00000498409.1 ENST00000475677.1 ENST00000478536.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr3_+_54157480 | 0.27 |
ENST00000490478.1
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr6_+_107349392 | 0.26 |
ENST00000443043.1
ENST00000405204.2 ENST00000311381.5 |
C6orf203
|
chromosome 6 open reading frame 203 |
| chr1_-_6557156 | 0.25 |
ENST00000537245.1
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr16_+_66995144 | 0.25 |
ENST00000394037.1
|
CES3
|
carboxylesterase 3 |
| chr19_-_48867171 | 0.25 |
ENST00000377431.2
ENST00000436660.2 ENST00000541566.1 |
TMEM143
|
transmembrane protein 143 |
| chr16_+_66995121 | 0.24 |
ENST00000303334.4
|
CES3
|
carboxylesterase 3 |
| chr3_-_28390581 | 0.24 |
ENST00000479665.1
|
AZI2
|
5-azacytidine induced 2 |
| chr22_+_31090793 | 0.24 |
ENST00000332585.6
ENST00000382310.3 ENST00000446658.2 |
OSBP2
|
oxysterol binding protein 2 |
| chr2_-_64371546 | 0.23 |
ENST00000358912.4
|
PELI1
|
pellino E3 ubiquitin protein ligase 1 |
| chr21_-_46515493 | 0.23 |
ENST00000600921.1
|
PRED57
|
HCG401283; PRED57 protein; Uncharacterized protein |
| chr10_-_119806085 | 0.23 |
ENST00000355624.3
|
RAB11FIP2
|
RAB11 family interacting protein 2 (class I) |
| chr3_+_183873098 | 0.22 |
ENST00000313143.3
|
DVL3
|
dishevelled segment polarity protein 3 |
| chr9_-_44402427 | 0.22 |
ENST00000540551.1
|
BX088651.1
|
LOC100126582 protein; Uncharacterized protein |
| chr19_-_51220176 | 0.22 |
ENST00000359082.3
ENST00000293441.1 |
SHANK1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr3_+_111260856 | 0.22 |
ENST00000352690.4
|
CD96
|
CD96 molecule |
| chr19_-_48867291 | 0.21 |
ENST00000435956.3
|
TMEM143
|
transmembrane protein 143 |
| chr3_-_38071122 | 0.21 |
ENST00000334661.4
|
PLCD1
|
phospholipase C, delta 1 |
| chr4_+_25915822 | 0.21 |
ENST00000506197.2
|
SMIM20
|
small integral membrane protein 20 |
| chr10_-_75634260 | 0.21 |
ENST00000372765.1
ENST00000351293.3 |
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr10_-_75634326 | 0.20 |
ENST00000322635.3
ENST00000444854.2 ENST00000423381.1 ENST00000322680.3 ENST00000394762.2 |
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr19_-_44860820 | 0.20 |
ENST00000354340.4
ENST00000337401.4 ENST00000587909.1 |
ZNF112
|
zinc finger protein 112 |
| chr3_+_122296443 | 0.20 |
ENST00000464300.2
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15 |
| chr17_-_74023474 | 0.20 |
ENST00000301607.3
|
EVPL
|
envoplakin |
| chr4_+_25915896 | 0.20 |
ENST00000514384.1
|
SMIM20
|
small integral membrane protein 20 |
| chr7_+_94139105 | 0.20 |
ENST00000297273.4
|
CASD1
|
CAS1 domain containing 1 |
| chr12_-_8088871 | 0.19 |
ENST00000075120.7
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr14_-_25103472 | 0.19 |
ENST00000216341.4
ENST00000382542.1 ENST00000382540.1 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr1_+_86046433 | 0.19 |
ENST00000451137.2
|
CYR61
|
cysteine-rich, angiogenic inducer, 61 |
| chr11_+_7273181 | 0.19 |
ENST00000318881.6
|
SYT9
|
synaptotagmin IX |
| chr14_-_25103388 | 0.18 |
ENST00000526004.1
ENST00000415355.3 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr12_+_58013693 | 0.18 |
ENST00000320442.4
ENST00000379218.2 |
SLC26A10
|
solute carrier family 26, member 10 |
| chr2_-_219433014 | 0.18 |
ENST00000418019.1
ENST00000454775.1 ENST00000338465.5 ENST00000415516.1 ENST00000258399.3 |
USP37
|
ubiquitin specific peptidase 37 |
| chr14_+_101297740 | 0.18 |
ENST00000555928.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr3_-_126327398 | 0.18 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr19_+_19496728 | 0.18 |
ENST00000537887.1
ENST00000417582.2 |
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr18_+_3449330 | 0.18 |
ENST00000549253.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr3_+_52489606 | 0.17 |
ENST00000488380.1
ENST00000420808.2 |
NISCH
|
nischarin |
| chr6_+_118228657 | 0.17 |
ENST00000360388.4
|
SLC35F1
|
solute carrier family 35, member F1 |
| chr4_-_171011084 | 0.17 |
ENST00000337664.4
|
AADAT
|
aminoadipate aminotransferase |
| chr6_-_49681235 | 0.17 |
ENST00000339139.4
|
CRISP2
|
cysteine-rich secretory protein 2 |
| chr3_-_57113314 | 0.17 |
ENST00000338458.4
ENST00000468727.1 |
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr4_-_171011323 | 0.17 |
ENST00000509167.1
ENST00000353187.2 ENST00000507375.1 ENST00000515480.1 |
AADAT
|
aminoadipate aminotransferase |
| chr9_+_74526384 | 0.16 |
ENST00000334731.2
ENST00000377031.3 |
C9orf85
|
chromosome 9 open reading frame 85 |
| chr20_+_62369623 | 0.16 |
ENST00000467211.1
|
RP4-583P15.14
|
RP4-583P15.14 |
| chr19_+_35629702 | 0.16 |
ENST00000351325.4
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr19_+_10563567 | 0.16 |
ENST00000344979.3
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr1_+_12123414 | 0.16 |
ENST00000263932.2
|
TNFRSF8
|
tumor necrosis factor receptor superfamily, member 8 |
| chr5_+_139927213 | 0.16 |
ENST00000310331.2
|
EIF4EBP3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr14_-_34931458 | 0.16 |
ENST00000298130.4
|
SPTSSA
|
serine palmitoyltransferase, small subunit A |
| chr5_+_137419581 | 0.15 |
ENST00000506684.1
ENST00000504809.1 ENST00000398754.1 |
WNT8A
|
wingless-type MMTV integration site family, member 8A |
| chr11_+_64073699 | 0.15 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr11_-_10879572 | 0.15 |
ENST00000413761.2
|
ZBED5
|
zinc finger, BED-type containing 5 |
| chr10_+_47083454 | 0.15 |
ENST00000374312.1
|
NPY4R
|
neuropeptide Y receptor Y4 |
| chrX_+_106449862 | 0.15 |
ENST00000372453.3
ENST00000535523.1 |
PIH1D3
|
PIH1 domain containing 3 |
| chr19_-_633576 | 0.15 |
ENST00000588649.2
|
POLRMT
|
polymerase (RNA) mitochondrial (DNA directed) |
| chr1_-_109584768 | 0.15 |
ENST00000357672.3
|
WDR47
|
WD repeat domain 47 |
| chr6_+_96025341 | 0.14 |
ENST00000369293.1
ENST00000358812.4 |
MANEA
|
mannosidase, endo-alpha |
| chr1_+_44412577 | 0.14 |
ENST00000372343.3
|
IPO13
|
importin 13 |
| chr1_-_109584716 | 0.14 |
ENST00000531337.1
ENST00000529074.1 ENST00000369965.4 |
WDR47
|
WD repeat domain 47 |
| chr1_+_25598989 | 0.14 |
ENST00000454452.2
|
RHD
|
Rh blood group, D antigen |
| chr20_-_3644046 | 0.14 |
ENST00000290417.2
ENST00000319242.3 |
GFRA4
|
GDNF family receptor alpha 4 |
| chr17_+_43318434 | 0.14 |
ENST00000587489.1
|
FMNL1
|
formin-like 1 |
| chr17_-_27038765 | 0.13 |
ENST00000581289.1
ENST00000301039.2 |
PROCA1
|
protein interacting with cyclin A1 |
| chr9_+_74526532 | 0.13 |
ENST00000486911.2
|
C9orf85
|
chromosome 9 open reading frame 85 |
| chr1_-_225616515 | 0.13 |
ENST00000338179.2
ENST00000425080.1 |
LBR
|
lamin B receptor |
| chr5_+_132208014 | 0.13 |
ENST00000296877.2
|
LEAP2
|
liver expressed antimicrobial peptide 2 |
| chrX_+_69454505 | 0.13 |
ENST00000374521.3
|
AWAT1
|
acyl-CoA wax alcohol acyltransferase 1 |
| chr6_+_127588020 | 0.13 |
ENST00000309649.3
ENST00000610162.1 ENST00000610153.1 ENST00000608991.1 ENST00000480444.1 |
RNF146
|
ring finger protein 146 |
| chr19_-_18385246 | 0.13 |
ENST00000608950.1
ENST00000600328.3 ENST00000600359.3 ENST00000392413.4 |
KIAA1683
|
KIAA1683 |
| chr3_+_52489503 | 0.13 |
ENST00000345716.4
|
NISCH
|
nischarin |
| chr17_-_43568062 | 0.12 |
ENST00000421073.2
ENST00000584420.1 ENST00000589780.1 ENST00000430334.3 |
PLEKHM1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 |
| chr17_-_73257667 | 0.12 |
ENST00000538886.1
ENST00000580799.1 ENST00000351904.7 ENST00000537686.1 |
GGA3
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 3 |
| chr11_-_10879593 | 0.12 |
ENST00000528289.1
ENST00000432999.2 |
ZBED5
|
zinc finger, BED-type containing 5 |
| chr1_-_16539094 | 0.12 |
ENST00000270747.3
|
ARHGEF19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chrX_+_66764375 | 0.12 |
ENST00000374690.3
|
AR
|
androgen receptor |
| chr3_-_50383096 | 0.12 |
ENST00000442887.1
ENST00000360165.3 |
ZMYND10
|
zinc finger, MYND-type containing 10 |
| chr22_+_20105012 | 0.12 |
ENST00000331821.3
ENST00000411892.1 |
RANBP1
|
RAN binding protein 1 |
| chr5_+_79615790 | 0.12 |
ENST00000296739.4
|
SPZ1
|
spermatogenic leucine zipper 1 |
| chr3_+_184055240 | 0.12 |
ENST00000383847.2
|
FAM131A
|
family with sequence similarity 131, member A |
| chr17_-_4852332 | 0.12 |
ENST00000572383.1
|
PFN1
|
profilin 1 |
| chr1_-_109584608 | 0.11 |
ENST00000400794.3
ENST00000528747.1 ENST00000369962.3 ENST00000361054.3 |
WDR47
|
WD repeat domain 47 |
| chr3_+_121903181 | 0.11 |
ENST00000498619.1
|
CASR
|
calcium-sensing receptor |
| chr15_+_83776137 | 0.11 |
ENST00000322019.9
|
TM6SF1
|
transmembrane 6 superfamily member 1 |
| chr3_+_160117418 | 0.11 |
ENST00000465903.1
ENST00000485645.1 ENST00000360111.2 ENST00000472991.1 ENST00000467468.1 ENST00000469762.1 ENST00000489573.1 ENST00000462787.1 ENST00000490207.1 ENST00000485867.1 |
SMC4
|
structural maintenance of chromosomes 4 |
| chr6_+_44126545 | 0.11 |
ENST00000532171.1
ENST00000398776.1 ENST00000542245.1 |
CAPN11
|
calpain 11 |
| chr6_+_127587755 | 0.11 |
ENST00000368314.1
ENST00000476956.1 ENST00000609447.1 ENST00000356799.2 ENST00000477776.1 ENST00000609944.1 |
RNF146
|
ring finger protein 146 |
| chr1_+_151483855 | 0.11 |
ENST00000427934.2
ENST00000271636.7 |
CGN
|
cingulin |
| chr3_-_126236605 | 0.11 |
ENST00000290868.2
|
UROC1
|
urocanate hydratase 1 |
| chrX_-_47930980 | 0.11 |
ENST00000442455.3
ENST00000428686.1 ENST00000276054.4 |
ZNF630
|
zinc finger protein 630 |
| chr6_+_152011628 | 0.11 |
ENST00000404742.1
ENST00000440973.1 |
ESR1
|
estrogen receptor 1 |
| chr3_+_160117087 | 0.10 |
ENST00000357388.3
|
SMC4
|
structural maintenance of chromosomes 4 |
| chr22_+_32149927 | 0.10 |
ENST00000437411.1
ENST00000535622.1 ENST00000536766.1 ENST00000400242.3 ENST00000266091.3 ENST00000400249.2 ENST00000400246.1 ENST00000382105.2 |
DEPDC5
|
DEP domain containing 5 |
| chr19_+_1026298 | 0.10 |
ENST00000263097.4
|
CNN2
|
calponin 2 |
| chr17_-_26879567 | 0.10 |
ENST00000581945.1
ENST00000444148.1 ENST00000301032.4 ENST00000335765.4 |
UNC119
|
unc-119 homolog (C. elegans) |
| chr16_-_67700594 | 0.10 |
ENST00000602644.1
ENST00000243878.4 |
ENKD1
|
enkurin domain containing 1 |
| chr2_-_182545603 | 0.09 |
ENST00000295108.3
|
NEUROD1
|
neuronal differentiation 1 |
| chr10_+_64564469 | 0.09 |
ENST00000373783.1
|
ADO
|
2-aminoethanethiol (cysteamine) dioxygenase |
| chr19_+_1026566 | 0.09 |
ENST00000348419.3
ENST00000565096.2 ENST00000562958.2 ENST00000562075.2 ENST00000607102.1 |
CNN2
|
calponin 2 |
| chr2_-_74374995 | 0.09 |
ENST00000295326.4
|
BOLA3
|
bolA family member 3 |
| chr19_-_36304201 | 0.09 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr3_+_183873240 | 0.09 |
ENST00000431765.1
|
DVL3
|
dishevelled segment polarity protein 3 |
| chr22_-_21579843 | 0.09 |
ENST00000405188.4
|
GGT2
|
gamma-glutamyltransferase 2 |
| chr7_+_49813255 | 0.08 |
ENST00000340652.4
|
VWC2
|
von Willebrand factor C domain containing 2 |
| chr10_-_35104185 | 0.08 |
ENST00000374789.3
ENST00000374788.3 ENST00000346874.4 ENST00000374794.3 ENST00000350537.4 ENST00000374790.3 ENST00000374776.1 ENST00000374773.1 ENST00000545693.1 ENST00000545260.1 ENST00000340077.5 |
PARD3
|
par-3 family cell polarity regulator |
| chr12_-_56727676 | 0.08 |
ENST00000547572.1
ENST00000257931.5 ENST00000440411.3 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr5_+_145316120 | 0.08 |
ENST00000359120.4
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr22_-_50700140 | 0.08 |
ENST00000215659.8
|
MAPK12
|
mitogen-activated protein kinase 12 |
| chr22_+_21321531 | 0.08 |
ENST00000405089.1
ENST00000335375.5 |
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3 |
| chr1_-_25747283 | 0.08 |
ENST00000346452.4
ENST00000340849.4 ENST00000349438.4 ENST00000294413.7 ENST00000413854.1 ENST00000455194.1 ENST00000243186.6 ENST00000425135.1 |
RHCE
|
Rh blood group, CcEe antigens |
| chr5_+_149569520 | 0.08 |
ENST00000230671.2
ENST00000524041.1 |
SLC6A7
|
solute carrier family 6 (neurotransmitter transporter), member 7 |
| chr17_-_73840415 | 0.08 |
ENST00000592386.1
ENST00000412096.2 ENST00000586147.1 |
UNC13D
|
unc-13 homolog D (C. elegans) |
| chrX_+_148793714 | 0.08 |
ENST00000355220.5
|
MAGEA11
|
melanoma antigen family A, 11 |
| chr19_+_10196981 | 0.07 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr19_+_1071203 | 0.07 |
ENST00000543365.1
|
HMHA1
|
histocompatibility (minor) HA-1 |
| chr10_+_112404132 | 0.07 |
ENST00000369519.3
|
RBM20
|
RNA binding motif protein 20 |
| chr13_+_27844464 | 0.07 |
ENST00000241463.4
|
RASL11A
|
RAS-like, family 11, member A |
| chr19_-_51054299 | 0.07 |
ENST00000599957.1
|
LRRC4B
|
leucine rich repeat containing 4B |
| chr3_+_52828805 | 0.07 |
ENST00000416872.2
ENST00000449956.2 |
ITIH3
|
inter-alpha-trypsin inhibitor heavy chain 3 |
| chr11_-_7041466 | 0.07 |
ENST00000536068.1
ENST00000278314.4 |
ZNF214
|
zinc finger protein 214 |
| chr3_+_111260954 | 0.07 |
ENST00000283285.5
|
CD96
|
CD96 molecule |
| chr14_-_24553834 | 0.07 |
ENST00000397002.2
|
NRL
|
neural retina leucine zipper |
| chr11_+_45868957 | 0.07 |
ENST00000443527.2
|
CRY2
|
cryptochrome 2 (photolyase-like) |
| chr5_-_177423243 | 0.06 |
ENST00000308304.2
|
PROP1
|
PROP paired-like homeobox 1 |
| chr14_+_24025462 | 0.06 |
ENST00000556015.1
ENST00000554970.1 ENST00000554789.1 |
THTPA
|
thiamine triphosphatase |
| chr16_-_69166460 | 0.06 |
ENST00000523421.1
ENST00000448552.2 ENST00000306585.6 ENST00000567763.1 ENST00000522497.1 ENST00000522091.1 ENST00000519520.1 |
CHTF8
|
CTF8, chromosome transmission fidelity factor 8 homolog (S. cerevisiae) |
| chr4_+_184427235 | 0.06 |
ENST00000412117.1
ENST00000434682.2 |
ING2
|
inhibitor of growth family, member 2 |
| chr14_+_24025194 | 0.06 |
ENST00000404535.3
ENST00000288014.6 |
THTPA
|
thiamine triphosphatase |
| chrX_+_47420516 | 0.06 |
ENST00000377045.4
ENST00000290277.6 ENST00000377039.2 |
ARAF
|
v-raf murine sarcoma 3611 viral oncogene homolog |
| chr3_-_58652523 | 0.06 |
ENST00000489857.1
ENST00000358781.2 |
FAM3D
|
family with sequence similarity 3, member D |
| chr3_-_128294929 | 0.06 |
ENST00000356020.2
|
C3orf27
|
chromosome 3 open reading frame 27 |
| chr13_-_38172863 | 0.06 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chr14_-_54908043 | 0.06 |
ENST00000556113.1
ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr3_-_113918254 | 0.06 |
ENST00000460779.1
|
DRD3
|
dopamine receptor D3 |
| chr17_-_4852243 | 0.06 |
ENST00000225655.5
|
PFN1
|
profilin 1 |
| chr15_-_75932528 | 0.06 |
ENST00000403490.1
|
IMP3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr1_+_32042105 | 0.06 |
ENST00000457433.2
ENST00000441210.2 |
TINAGL1
|
tubulointerstitial nephritis antigen-like 1 |
| chr22_+_20104947 | 0.05 |
ENST00000402752.1
|
RANBP1
|
RAN binding protein 1 |
| chr12_-_56727487 | 0.05 |
ENST00000548043.1
ENST00000425394.2 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr19_+_6361841 | 0.05 |
ENST00000596605.1
|
CLPP
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr4_+_152330390 | 0.05 |
ENST00000503146.1
ENST00000435205.1 |
FAM160A1
|
family with sequence similarity 160, member A1 |
| chr19_-_11689752 | 0.05 |
ENST00000592659.1
ENST00000592828.1 ENST00000218758.5 ENST00000412435.2 |
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr1_+_32042131 | 0.05 |
ENST00000271064.7
ENST00000537531.1 |
TINAGL1
|
tubulointerstitial nephritis antigen-like 1 |
| chr11_-_33795893 | 0.05 |
ENST00000526785.1
ENST00000534136.1 ENST00000265651.3 ENST00000530401.1 ENST00000448981.2 |
FBXO3
|
F-box protein 3 |
| chr3_-_28390415 | 0.05 |
ENST00000414162.1
ENST00000420543.2 |
AZI2
|
5-azacytidine induced 2 |
| chr1_+_25599018 | 0.05 |
ENST00000417538.2
ENST00000357542.4 ENST00000568195.1 ENST00000342055.5 ENST00000423810.2 |
RHD
|
Rh blood group, D antigen |
| chr2_-_74780176 | 0.05 |
ENST00000409549.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr14_+_22356029 | 0.05 |
ENST00000390437.2
|
TRAV12-2
|
T cell receptor alpha variable 12-2 |
| chr8_-_131028660 | 0.05 |
ENST00000401979.2
ENST00000517654.1 ENST00000522361.1 ENST00000518167.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr3_+_122296465 | 0.05 |
ENST00000483793.1
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15 |
| chr7_+_99102573 | 0.05 |
ENST00000394170.2
|
ZKSCAN5
|
zinc finger with KRAB and SCAN domains 5 |
| chr7_-_752074 | 0.05 |
ENST00000360274.4
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr2_+_108905325 | 0.05 |
ENST00000438339.1
ENST00000409880.1 ENST00000437390.2 |
SULT1C2
|
sulfotransferase family, cytosolic, 1C, member 2 |
| chr5_-_33892046 | 0.05 |
ENST00000352040.3
|
ADAMTS12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr12_-_58135903 | 0.05 |
ENST00000257897.3
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr6_-_33771757 | 0.04 |
ENST00000507738.1
ENST00000266003.5 ENST00000430124.2 |
MLN
|
motilin |
| chr17_-_73975444 | 0.04 |
ENST00000293217.5
ENST00000537812.1 |
ACOX1
|
acyl-CoA oxidase 1, palmitoyl |
| chr11_-_62380199 | 0.04 |
ENST00000419857.1
ENST00000394773.2 |
EML3
|
echinoderm microtubule associated protein like 3 |
| chr8_-_131028869 | 0.04 |
ENST00000518283.1
ENST00000519110.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr2_+_179345173 | 0.04 |
ENST00000234453.5
|
PLEKHA3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 |
| chr3_-_15540055 | 0.04 |
ENST00000605797.1
ENST00000435459.2 |
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr16_-_30537839 | 0.04 |
ENST00000380412.5
|
ZNF768
|
zinc finger protein 768 |
| chr1_-_16939976 | 0.04 |
ENST00000430580.2
|
NBPF1
|
neuroblastoma breakpoint family, member 1 |
| chrX_-_106449656 | 0.04 |
ENST00000372466.4
ENST00000421752.1 ENST00000372461.3 |
NUP62CL
|
nucleoporin 62kDa C-terminal like |
| chr22_+_30163340 | 0.04 |
ENST00000330029.6
ENST00000401406.3 |
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr1_+_29241027 | 0.04 |
ENST00000373797.1
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr1_+_178482262 | 0.04 |
ENST00000367641.3
ENST00000367639.1 |
TEX35
|
testis expressed 35 |
| chr5_+_139944396 | 0.04 |
ENST00000514199.1
|
SLC35A4
|
solute carrier family 35, member A4 |
| chr5_+_139944024 | 0.04 |
ENST00000323146.3
|
SLC35A4
|
solute carrier family 35, member A4 |
| chr4_+_123073464 | 0.04 |
ENST00000264501.4
|
KIAA1109
|
KIAA1109 |
| chrX_+_7137475 | 0.04 |
ENST00000217961.4
|
STS
|
steroid sulfatase (microsomal), isozyme S |
| chr22_-_30234218 | 0.04 |
ENST00000307790.3
ENST00000542393.1 ENST00000397771.2 |
ASCC2
|
activating signal cointegrator 1 complex subunit 2 |
| chr22_+_20105259 | 0.04 |
ENST00000416427.1
ENST00000421656.1 ENST00000423859.1 ENST00000418705.2 |
RANBP1
|
RAN binding protein 1 |
| chr3_-_49158218 | 0.03 |
ENST00000417901.1
ENST00000306026.5 ENST00000434032.2 |
USP19
|
ubiquitin specific peptidase 19 |
| chr3_-_28390298 | 0.03 |
ENST00000457172.1
|
AZI2
|
5-azacytidine induced 2 |
| chr2_+_108905095 | 0.03 |
ENST00000251481.6
ENST00000326853.5 |
SULT1C2
|
sulfotransferase family, cytosolic, 1C, member 2 |
| chr3_-_194119083 | 0.03 |
ENST00000401815.1
|
GP5
|
glycoprotein V (platelet) |
| chr5_-_83680603 | 0.03 |
ENST00000296591.5
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1H4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 0.3 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.1 | 0.5 | GO:2000015 | negative regulation of CREB transcription factor activity(GO:0032792) regulation of determination of dorsal identity(GO:2000015) |
| 0.1 | 0.3 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.1 | 0.2 | GO:0061316 | canonical Wnt signaling pathway involved in heart development(GO:0061316) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) |
| 0.1 | 0.2 | GO:0050894 | determination of affect(GO:0050894) |
| 0.1 | 0.4 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.1 | 0.2 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.1 | 0.3 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.6 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 0.3 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 0.4 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.1 | 0.4 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 0.2 | GO:0032639 | TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.1 | 0.3 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.2 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0060769 | male genitalia morphogenesis(GO:0048808) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.6 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.2 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.3 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.3 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:2001287 | negative regulation of clathrin-mediated endocytosis(GO:1900186) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.2 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.0 | 0.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.1 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.1 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.1 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.4 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.4 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 | 0.3 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.4 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.1 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.0 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.3 | GO:0048243 | norepinephrine secretion(GO:0048243) |
| 0.0 | 0.1 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.1 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.1 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.0 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) bone regeneration(GO:1990523) |
| 0.0 | 0.1 | GO:0060737 | prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.2 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.4 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.2 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.3 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.2 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.0 | 0.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.5 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.7 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.4 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.2 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.1 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.4 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |