Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
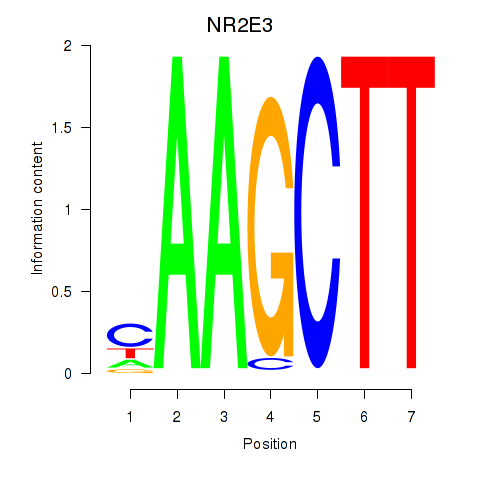
Results for NR2E3
Z-value: 1.19
Transcription factors associated with NR2E3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2E3
|
ENSG00000031544.10 | nuclear receptor subfamily 2 group E member 3 |
Activity profile of NR2E3 motif
Sorted Z-values of NR2E3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_61547894 | 7.62 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
| chr1_+_61547405 | 6.40 |
ENST00000371189.4
|
NFIA
|
nuclear factor I/A |
| chr1_+_61330931 | 4.88 |
ENST00000371191.1
|
NFIA
|
nuclear factor I/A |
| chr9_+_72002837 | 4.36 |
ENST00000377216.3
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr12_+_32655048 | 3.94 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr17_+_68165657 | 3.67 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr11_-_33913708 | 3.66 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr7_+_94023873 | 3.44 |
ENST00000297268.6
|
COL1A2
|
collagen, type I, alpha 2 |
| chr12_+_79258444 | 3.13 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr6_-_76203345 | 3.04 |
ENST00000393004.2
|
FILIP1
|
filamin A interacting protein 1 |
| chr3_-_64211112 | 2.98 |
ENST00000295902.6
|
PRICKLE2
|
prickle homolog 2 (Drosophila) |
| chr12_+_79258547 | 2.98 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr18_+_8717369 | 2.87 |
ENST00000359865.3
ENST00000400050.3 ENST00000306285.7 |
SOGA2
|
SOGA family member 2 |
| chr12_+_93963590 | 2.81 |
ENST00000340600.2
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr4_-_186877806 | 2.80 |
ENST00000355634.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr6_-_76203454 | 2.69 |
ENST00000237172.7
|
FILIP1
|
filamin A interacting protein 1 |
| chr1_+_84630053 | 2.53 |
ENST00000394838.2
ENST00000370682.3 ENST00000432111.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr1_+_164528866 | 2.48 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr18_+_42260861 | 2.48 |
ENST00000282030.5
|
SETBP1
|
SET binding protein 1 |
| chr1_+_84630645 | 2.37 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr12_+_53443963 | 2.30 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr13_-_45010939 | 2.29 |
ENST00000261489.2
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr5_+_68788594 | 2.23 |
ENST00000396442.2
ENST00000380766.2 |
OCLN
|
occludin |
| chr20_+_42574317 | 2.22 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2 |
| chr5_-_138210977 | 2.17 |
ENST00000274711.6
ENST00000521094.2 |
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr1_+_221051699 | 2.12 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr9_+_71986182 | 2.10 |
ENST00000303068.7
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr21_+_17792672 | 2.07 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr3_+_193853927 | 2.03 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr10_-_93392811 | 1.96 |
ENST00000238994.5
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C |
| chr19_-_14628645 | 1.95 |
ENST00000598235.1
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr1_+_87797351 | 1.91 |
ENST00000370542.1
|
LMO4
|
LIM domain only 4 |
| chr9_-_14180778 | 1.90 |
ENST00000380924.1
ENST00000543693.1 |
NFIB
|
nuclear factor I/B |
| chr9_+_139606983 | 1.87 |
ENST00000371692.4
|
FAM69B
|
family with sequence similarity 69, member B |
| chr18_-_53069419 | 1.78 |
ENST00000570177.2
|
TCF4
|
transcription factor 4 |
| chr3_-_169381183 | 1.78 |
ENST00000494292.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr12_+_56661461 | 1.76 |
ENST00000546544.1
ENST00000553234.1 |
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae) |
| chr1_+_66999799 | 1.75 |
ENST00000371035.3
ENST00000371036.3 ENST00000371037.4 |
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr2_+_110656005 | 1.74 |
ENST00000437679.2
|
LIMS3
|
LIM and senescent cell antigen-like domains 3 |
| chr9_+_71939488 | 1.68 |
ENST00000455972.1
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chrX_-_117119243 | 1.66 |
ENST00000539496.1
ENST00000469946.1 |
KLHL13
|
kelch-like family member 13 |
| chr9_+_71944241 | 1.66 |
ENST00000257515.8
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr7_+_106809406 | 1.62 |
ENST00000468410.1
ENST00000478930.1 ENST00000464009.1 ENST00000222574.4 |
HBP1
|
HMG-box transcription factor 1 |
| chr7_+_102553430 | 1.62 |
ENST00000339431.4
ENST00000249377.4 |
LRRC17
|
leucine rich repeat containing 17 |
| chr1_+_89829610 | 1.60 |
ENST00000370456.4
ENST00000535065.1 |
GBP6
|
guanylate binding protein family, member 6 |
| chr2_+_33359646 | 1.59 |
ENST00000390003.4
ENST00000418533.2 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr20_-_45981138 | 1.58 |
ENST00000446994.2
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr2_+_33359687 | 1.57 |
ENST00000402934.1
ENST00000404525.1 ENST00000407925.1 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr1_+_65775204 | 1.54 |
ENST00000371069.4
|
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr3_-_65583561 | 1.53 |
ENST00000460329.2
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr11_+_121447469 | 1.47 |
ENST00000532694.1
ENST00000534286.1 |
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr2_+_173600565 | 1.46 |
ENST00000397081.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr2_-_3584430 | 1.42 |
ENST00000438482.1
ENST00000422961.1 |
AC108488.4
|
AC108488.4 |
| chr12_+_56661033 | 1.35 |
ENST00000433805.2
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae) |
| chr15_-_52587945 | 1.33 |
ENST00000443683.2
ENST00000558479.1 ENST00000261839.7 |
MYO5C
|
myosin VC |
| chr19_-_14629224 | 1.31 |
ENST00000254322.2
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr5_-_90679145 | 1.31 |
ENST00000265138.3
|
ARRDC3
|
arrestin domain containing 3 |
| chr2_+_173600514 | 1.31 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr4_-_25865159 | 1.30 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chrX_-_70474910 | 1.24 |
ENST00000373988.1
ENST00000373998.1 |
ZMYM3
|
zinc finger, MYM-type 3 |
| chr15_+_65914260 | 1.24 |
ENST00000261892.6
ENST00000339868.6 |
SLC24A1
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 |
| chr11_+_92085707 | 1.24 |
ENST00000525166.1
|
FAT3
|
FAT atypical cadherin 3 |
| chr7_-_121784285 | 1.21 |
ENST00000417368.2
|
AASS
|
aminoadipate-semialdehyde synthase |
| chr6_+_74405501 | 1.21 |
ENST00000437994.2
ENST00000422508.2 |
CD109
|
CD109 molecule |
| chr1_-_153599426 | 1.20 |
ENST00000392622.1
|
S100A13
|
S100 calcium binding protein A13 |
| chr2_+_190722119 | 1.19 |
ENST00000452382.1
|
PMS1
|
PMS1 postmeiotic segregation increased 1 (S. cerevisiae) |
| chr20_-_17539456 | 1.16 |
ENST00000544874.1
ENST00000377868.2 |
BFSP1
|
beaded filament structural protein 1, filensin |
| chrX_+_102883887 | 1.14 |
ENST00000372625.3
ENST00000372624.3 |
TCEAL1
|
transcription elongation factor A (SII)-like 1 |
| chr12_-_10541575 | 1.10 |
ENST00000540818.1
|
KLRK1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr1_-_43282906 | 1.08 |
ENST00000372521.4
|
CCDC23
|
coiled-coil domain containing 23 |
| chr2_+_173686303 | 1.08 |
ENST00000397087.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr2_-_158732340 | 1.06 |
ENST00000539637.1
ENST00000413751.1 ENST00000434821.1 ENST00000424669.1 |
ACVR1
|
activin A receptor, type I |
| chr3_-_129407535 | 1.05 |
ENST00000432054.2
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chrX_-_102565858 | 1.04 |
ENST00000449185.1
ENST00000536889.1 |
BEX2
|
brain expressed X-linked 2 |
| chr3_-_165555200 | 1.04 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr1_+_66999268 | 1.04 |
ENST00000371039.1
ENST00000424320.1 |
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr4_+_174089904 | 1.02 |
ENST00000265000.4
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr22_-_37915247 | 1.01 |
ENST00000251973.5
|
CARD10
|
caspase recruitment domain family, member 10 |
| chr6_-_42016385 | 1.01 |
ENST00000502771.1
ENST00000508143.1 ENST00000514588.1 ENST00000510503.1 ENST00000415497.2 ENST00000372988.4 |
CCND3
|
cyclin D3 |
| chr19_-_9546227 | 1.00 |
ENST00000361451.2
ENST00000361151.1 |
ZNF266
|
zinc finger protein 266 |
| chr4_-_110723335 | 0.99 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chr4_-_110723194 | 0.99 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr21_-_35016231 | 0.97 |
ENST00000438788.1
|
CRYZL1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr12_+_48516357 | 0.97 |
ENST00000549022.1
ENST00000547587.1 ENST00000312352.7 |
PFKM
|
phosphofructokinase, muscle |
| chr5_+_110559784 | 0.97 |
ENST00000282356.4
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV |
| chr9_+_134065506 | 0.97 |
ENST00000483497.2
|
NUP214
|
nucleoporin 214kDa |
| chr5_+_140571902 | 0.97 |
ENST00000239446.4
|
PCDHB10
|
protocadherin beta 10 |
| chr13_+_76378357 | 0.96 |
ENST00000489941.2
ENST00000525373.1 |
LMO7
|
LIM domain 7 |
| chr11_-_82745238 | 0.96 |
ENST00000531021.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr15_+_67841330 | 0.96 |
ENST00000354498.5
|
MAP2K5
|
mitogen-activated protein kinase kinase 5 |
| chr19_-_54693401 | 0.95 |
ENST00000338624.6
|
MBOAT7
|
membrane bound O-acyltransferase domain containing 7 |
| chr3_+_141105235 | 0.95 |
ENST00000503809.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr1_-_153599732 | 0.94 |
ENST00000392623.1
|
S100A13
|
S100 calcium binding protein A13 |
| chr17_-_39150385 | 0.94 |
ENST00000391586.1
|
KRTAP3-3
|
keratin associated protein 3-3 |
| chr4_+_170581213 | 0.94 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr4_-_110723134 | 0.94 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr13_+_60971080 | 0.94 |
ENST00000377894.2
|
TDRD3
|
tudor domain containing 3 |
| chr3_-_197300194 | 0.92 |
ENST00000358186.2
ENST00000431056.1 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr2_-_45795145 | 0.92 |
ENST00000535761.1
|
SRBD1
|
S1 RNA binding domain 1 |
| chr8_-_6836156 | 0.91 |
ENST00000382679.2
|
DEFA1
|
defensin, alpha 1 |
| chr1_+_203595689 | 0.90 |
ENST00000357681.5
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr17_-_46691990 | 0.90 |
ENST00000576562.1
|
HOXB8
|
homeobox B8 |
| chr15_-_65067773 | 0.89 |
ENST00000300069.4
|
RBPMS2
|
RNA binding protein with multiple splicing 2 |
| chr11_+_92085262 | 0.87 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr15_+_75639296 | 0.86 |
ENST00000564500.1
ENST00000355059.4 ENST00000566752.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr1_+_43291220 | 0.85 |
ENST00000372514.3
|
ERMAP
|
erythroblast membrane-associated protein (Scianna blood group) |
| chr17_-_64188177 | 0.84 |
ENST00000535342.2
|
CEP112
|
centrosomal protein 112kDa |
| chr19_-_31840438 | 0.83 |
ENST00000240587.4
|
TSHZ3
|
teashirt zinc finger homeobox 3 |
| chr10_+_82168240 | 0.81 |
ENST00000372187.5
ENST00000372185.1 |
FAM213A
|
family with sequence similarity 213, member A |
| chr4_-_6711558 | 0.80 |
ENST00000320848.6
|
MRFAP1L1
|
Morf4 family associated protein 1-like 1 |
| chr12_+_96588279 | 0.80 |
ENST00000552142.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr8_+_74903580 | 0.80 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr2_-_216257849 | 0.79 |
ENST00000456923.1
|
FN1
|
fibronectin 1 |
| chr1_+_46049706 | 0.79 |
ENST00000527470.1
ENST00000525515.1 ENST00000537798.1 ENST00000402363.3 ENST00000528238.1 ENST00000350030.3 ENST00000470768.1 ENST00000372052.4 ENST00000351223.3 |
NASP
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr16_+_31404624 | 0.78 |
ENST00000389202.2
|
ITGAD
|
integrin, alpha D |
| chr2_+_24150180 | 0.77 |
ENST00000404924.1
|
UBXN2A
|
UBX domain protein 2A |
| chr1_-_47131521 | 0.77 |
ENST00000542495.1
ENST00000532925.1 |
ATPAF1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr19_+_17337473 | 0.77 |
ENST00000598068.1
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr6_+_130339710 | 0.76 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr1_+_84630367 | 0.76 |
ENST00000370680.1
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr8_+_103563792 | 0.76 |
ENST00000285402.3
|
ODF1
|
outer dense fiber of sperm tails 1 |
| chrX_+_17755563 | 0.73 |
ENST00000380045.3
ENST00000380041.3 ENST00000380043.3 ENST00000398080.1 |
SCML1
|
sex comb on midleg-like 1 (Drosophila) |
| chr20_-_56286479 | 0.73 |
ENST00000265626.4
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr1_-_8483723 | 0.73 |
ENST00000476556.1
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats |
| chr4_+_71588372 | 0.73 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr17_-_76870126 | 0.71 |
ENST00000586057.1
|
TIMP2
|
TIMP metallopeptidase inhibitor 2 |
| chr2_-_65593784 | 0.71 |
ENST00000443619.2
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chrX_-_10588595 | 0.71 |
ENST00000423614.1
ENST00000317552.4 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr11_+_86013253 | 0.71 |
ENST00000533986.1
ENST00000278483.3 |
C11orf73
|
chromosome 11 open reading frame 73 |
| chrX_+_37639264 | 0.70 |
ENST00000378588.4
|
CYBB
|
cytochrome b-245, beta polypeptide |
| chrX_-_10588459 | 0.70 |
ENST00000380782.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr4_-_39367949 | 0.70 |
ENST00000503784.1
ENST00000349703.2 ENST00000381897.1 |
RFC1
|
replication factor C (activator 1) 1, 145kDa |
| chr11_+_20385231 | 0.69 |
ENST00000530266.1
ENST00000421577.2 ENST00000443524.2 ENST00000419348.2 |
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa |
| chr18_+_7754957 | 0.69 |
ENST00000400053.4
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr4_+_76481258 | 0.69 |
ENST00000311623.4
ENST00000435974.2 |
C4orf26
|
chromosome 4 open reading frame 26 |
| chr3_+_113616317 | 0.69 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr12_-_51664058 | 0.69 |
ENST00000605627.1
|
SMAGP
|
small cell adhesion glycoprotein |
| chr4_-_152147579 | 0.69 |
ENST00000304527.4
ENST00000455740.1 ENST00000424281.1 ENST00000409598.4 |
SH3D19
|
SH3 domain containing 19 |
| chr18_-_52989217 | 0.68 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr1_-_17307173 | 0.68 |
ENST00000438542.1
ENST00000375535.3 |
MFAP2
|
microfibrillar-associated protein 2 |
| chr14_+_101297740 | 0.67 |
ENST00000555928.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr14_+_24584508 | 0.67 |
ENST00000559354.1
ENST00000560459.1 ENST00000559593.1 ENST00000396941.4 ENST00000396936.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr6_-_79944336 | 0.67 |
ENST00000344726.5
ENST00000275036.7 |
HMGN3
|
high mobility group nucleosomal binding domain 3 |
| chr8_-_82395461 | 0.67 |
ENST00000256104.4
|
FABP4
|
fatty acid binding protein 4, adipocyte |
| chr16_+_29819372 | 0.65 |
ENST00000568544.1
ENST00000569978.1 |
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr4_+_71587669 | 0.65 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr17_-_62208169 | 0.65 |
ENST00000606895.1
|
ERN1
|
endoplasmic reticulum to nucleus signaling 1 |
| chrX_-_24665208 | 0.64 |
ENST00000356768.4
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chrX_+_107683096 | 0.63 |
ENST00000328300.6
ENST00000361603.2 |
COL4A5
|
collagen, type IV, alpha 5 |
| chr3_-_125775629 | 0.63 |
ENST00000383598.2
|
SLC41A3
|
solute carrier family 41, member 3 |
| chr1_+_168250194 | 0.63 |
ENST00000367821.3
|
TBX19
|
T-box 19 |
| chr12_+_80603233 | 0.62 |
ENST00000547103.1
ENST00000458043.2 |
OTOGL
|
otogelin-like |
| chr17_-_40575535 | 0.62 |
ENST00000357037.5
|
PTRF
|
polymerase I and transcript release factor |
| chr19_-_9546177 | 0.61 |
ENST00000592292.1
ENST00000588221.1 |
ZNF266
|
zinc finger protein 266 |
| chr3_+_141043050 | 0.61 |
ENST00000509842.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr10_+_115939008 | 0.61 |
ENST00000369282.1
ENST00000251864.2 ENST00000369281.2 ENST00000422662.1 |
TDRD1
|
tudor domain containing 1 |
| chr16_+_29819446 | 0.60 |
ENST00000568282.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr19_+_47421933 | 0.60 |
ENST00000404338.3
|
ARHGAP35
|
Rho GTPase activating protein 35 |
| chr1_+_43282782 | 0.59 |
ENST00000372517.2
|
ERMAP
|
erythroblast membrane-associated protein (Scianna blood group) |
| chr10_-_50396407 | 0.59 |
ENST00000374153.2
ENST00000374151.3 |
C10orf128
|
chromosome 10 open reading frame 128 |
| chrX_+_54835493 | 0.56 |
ENST00000396224.1
|
MAGED2
|
melanoma antigen family D, 2 |
| chr19_-_37697976 | 0.56 |
ENST00000588873.1
|
CTC-454I21.3
|
Uncharacterized protein; Zinc finger protein 585B |
| chr21_-_27542972 | 0.56 |
ENST00000346798.3
ENST00000439274.2 ENST00000354192.3 ENST00000348990.5 ENST00000357903.3 ENST00000358918.3 ENST00000359726.3 |
APP
|
amyloid beta (A4) precursor protein |
| chr1_+_22963158 | 0.55 |
ENST00000438241.1
|
C1QA
|
complement component 1, q subcomponent, A chain |
| chr3_-_138312971 | 0.55 |
ENST00000485115.1
ENST00000484888.1 ENST00000468900.1 ENST00000542237.1 ENST00000481834.1 |
CEP70
|
centrosomal protein 70kDa |
| chr12_+_53773944 | 0.54 |
ENST00000551969.1
ENST00000327443.4 |
SP1
|
Sp1 transcription factor |
| chr14_+_54976603 | 0.54 |
ENST00000557317.1
|
CGRRF1
|
cell growth regulator with ring finger domain 1 |
| chr7_-_130080977 | 0.53 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr15_-_77197620 | 0.53 |
ENST00000565970.1
ENST00000563290.1 ENST00000565372.1 ENST00000564177.1 ENST00000568382.1 ENST00000563919.1 |
SCAPER
|
S-phase cyclin A-associated protein in the ER |
| chr14_+_22508822 | 0.53 |
ENST00000390448.3
|
TRAV20
|
T cell receptor alpha variable 20 |
| chr11_+_63997750 | 0.53 |
ENST00000321685.3
|
DNAJC4
|
DnaJ (Hsp40) homolog, subfamily C, member 4 |
| chr3_+_69915385 | 0.53 |
ENST00000314589.5
|
MITF
|
microphthalmia-associated transcription factor |
| chr4_+_159727272 | 0.52 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chr10_-_49459800 | 0.52 |
ENST00000305531.3
|
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr3_+_52811596 | 0.51 |
ENST00000542827.1
ENST00000273283.2 |
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr6_-_28220002 | 0.51 |
ENST00000377294.2
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4 |
| chr12_-_8043736 | 0.51 |
ENST00000539924.1
|
SLC2A14
|
solute carrier family 2 (facilitated glucose transporter), member 14 |
| chr16_-_15149828 | 0.50 |
ENST00000566419.1
ENST00000568320.1 |
NTAN1
|
N-terminal asparagine amidase |
| chr10_-_45474237 | 0.50 |
ENST00000448778.1
ENST00000298295.3 |
C10orf10
|
chromosome 10 open reading frame 10 |
| chr3_+_37035263 | 0.50 |
ENST00000458205.2
ENST00000539477.1 |
MLH1
|
mutL homolog 1 |
| chr11_-_5271122 | 0.50 |
ENST00000330597.3
|
HBG1
|
hemoglobin, gamma A |
| chr5_+_41925325 | 0.49 |
ENST00000296812.2
ENST00000281623.3 ENST00000509134.1 |
FBXO4
|
F-box protein 4 |
| chr3_-_196159268 | 0.49 |
ENST00000381887.3
ENST00000535858.1 ENST00000428095.1 ENST00000296328.4 |
UBXN7
|
UBX domain protein 7 |
| chr11_+_20385327 | 0.49 |
ENST00000451739.2
ENST00000532505.1 |
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa |
| chr19_+_6373715 | 0.49 |
ENST00000599849.1
|
ALKBH7
|
alkB, alkylation repair homolog 7 (E. coli) |
| chr1_+_203595903 | 0.49 |
ENST00000367218.3
ENST00000367219.3 ENST00000391954.2 |
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr6_+_42896865 | 0.48 |
ENST00000372836.4
ENST00000394142.3 |
CNPY3
|
canopy FGF signaling regulator 3 |
| chr17_+_45608430 | 0.48 |
ENST00000322157.4
|
NPEPPS
|
aminopeptidase puromycin sensitive |
| chr3_+_148508845 | 0.48 |
ENST00000491148.1
|
CPB1
|
carboxypeptidase B1 (tissue) |
| chr15_+_23255242 | 0.48 |
ENST00000450802.3
|
GOLGA8I
|
golgin A8 family, member I |
| chr15_+_75639951 | 0.48 |
ENST00000564784.1
ENST00000569035.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr2_-_201729393 | 0.46 |
ENST00000321356.4
|
CLK1
|
CDC-like kinase 1 |
| chr4_+_118955500 | 0.46 |
ENST00000296499.5
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr14_+_69658480 | 0.46 |
ENST00000409949.1
ENST00000409242.1 ENST00000312994.5 ENST00000413191.1 |
EXD2
|
exonuclease 3'-5' domain containing 2 |
| chr15_+_42696992 | 0.45 |
ENST00000561817.1
|
CAPN3
|
calpain 3, (p94) |
| chrX_-_135338503 | 0.45 |
ENST00000370663.5
|
MAP7D3
|
MAP7 domain containing 3 |
| chr1_+_24742264 | 0.45 |
ENST00000374399.4
ENST00000003912.3 ENST00000358028.4 ENST00000339255.2 |
NIPAL3
|
NIPA-like domain containing 3 |
| chr6_+_56911336 | 0.45 |
ENST00000370733.4
|
KIAA1586
|
KIAA1586 |
| chr1_-_8585945 | 0.44 |
ENST00000377464.1
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats |
| chr19_+_37808778 | 0.44 |
ENST00000544914.1
ENST00000589188.1 |
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2E3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.9 | 3.7 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.8 | 5.7 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.7 | 2.1 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.6 | 16.8 | GO:0072189 | ureter development(GO:0072189) |
| 0.5 | 1.5 | GO:1902960 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.5 | 1.4 | GO:0033241 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.4 | 1.3 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.4 | 1.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.4 | 1.1 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.3 | 1.0 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.3 | 1.3 | GO:0018032 | protein amidation(GO:0018032) |
| 0.3 | 1.0 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.3 | 3.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.3 | 1.2 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.3 | 0.9 | GO:0035915 | pore formation in membrane of other organism(GO:0035915) |
| 0.3 | 1.2 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.3 | 2.8 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.3 | 1.9 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.3 | 2.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.3 | 1.6 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.3 | 0.8 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.2 | 1.0 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.2 | 3.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.2 | 1.6 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.2 | 3.4 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.2 | 0.8 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.2 | 1.2 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.2 | 0.6 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.2 | 1.0 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.2 | 0.9 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.2 | 1.3 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.2 | 0.5 | GO:0036166 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.2 | 1.5 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.2 | 0.5 | GO:0051257 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.2 | 0.9 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.2 | 0.6 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.2 | 2.2 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.2 | 0.8 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 3.7 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.1 | 0.7 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.1 | 0.4 | GO:0035905 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.1 | 1.9 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) regulation of cell fate specification(GO:0042659) |
| 0.1 | 2.8 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 3.1 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 2.2 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.1 | 0.8 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.1 | 0.3 | GO:0098759 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.1 | 0.3 | GO:0045925 | female courtship behavior(GO:0008050) positive regulation of female receptivity(GO:0045925) |
| 0.1 | 0.3 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 0.4 | GO:0032053 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 1.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.5 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.1 | 0.7 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.1 | 0.9 | GO:1901142 | insulin metabolic process(GO:1901142) |
| 0.1 | 0.9 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 0.4 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 2.4 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.1 | 0.6 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.1 | 0.6 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.7 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 1.3 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.1 | 0.9 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 0.5 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.8 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 1.2 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 2.5 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.8 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.1 | 0.4 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.1 | 0.2 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 1.5 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.1 | 0.8 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.3 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.1 | 0.4 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.6 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.2 | GO:1904604 | regulation of connective tissue replacement involved in inflammatory response wound healing(GO:1904596) negative regulation of connective tissue replacement involved in inflammatory response wound healing(GO:1904597) regulation of advanced glycation end-product receptor activity(GO:1904603) negative regulation of advanced glycation end-product receptor activity(GO:1904604) negative regulation of connective tissue replacement(GO:1905204) |
| 0.1 | 0.8 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.1 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.1 | 0.8 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.1 | 0.6 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.6 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.2 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.1 | 0.4 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.1 | 1.7 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 0.4 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.5 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.8 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.7 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.4 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.5 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.1 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.7 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.7 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 1.5 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.2 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.5 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.0 | 0.7 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.7 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.2 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.0 | 0.3 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 1.0 | GO:1901661 | quinone metabolic process(GO:1901661) |
| 0.0 | 2.4 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.2 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 1.4 | GO:1904031 | positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.3 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.3 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.6 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 4.0 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 2.5 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.3 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 1.2 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.3 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.3 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.1 | GO:0046502 | uroporphyrinogen III biosynthetic process(GO:0006780) uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.7 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.7 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.4 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.4 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.2 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.1 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.0 | 0.3 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 1.0 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.1 | GO:0070197 | telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 2.3 | GO:0090175 | Wnt signaling pathway, planar cell polarity pathway(GO:0060071) regulation of establishment of planar polarity(GO:0090175) |
| 0.0 | 0.2 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.3 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.1 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.5 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.2 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.5 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.5 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 1.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.0 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.3 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.1 | GO:1904382 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.0 | 2.9 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.2 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.0 | 0.1 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.2 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 0.9 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.7 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.5 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.4 | 1.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.3 | 1.0 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.3 | 5.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 3.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 3.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 1.7 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.2 | 1.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.2 | 0.8 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 1.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 3.8 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.7 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.6 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.1 | 2.8 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 1.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.7 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.7 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 0.3 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 3.8 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.2 | GO:0097059 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.1 | 0.4 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 0.8 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.6 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 1.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.3 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 0.3 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.1 | 0.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.4 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.4 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 1.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.8 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.3 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.1 | GO:0039713 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 1.4 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 1.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.4 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.4 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.5 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 1.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.9 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 5.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.1 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.0 | 0.3 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 1.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.3 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 1.0 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.0 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.8 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.7 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 1.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 2.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.7 | GO:0031901 | early endosome membrane(GO:0031901) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 6.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.6 | 3.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.5 | 2.8 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.5 | 1.4 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.4 | 1.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.4 | 1.2 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.3 | 3.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.3 | 1.3 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.3 | 5.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.3 | 0.9 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.3 | 1.0 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.2 | 0.7 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 3.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 1.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 1.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.2 | 3.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 2.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 0.5 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.2 | 1.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 0.9 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 0.6 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.1 | 0.4 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 0.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.3 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 0.3 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.1 | 1.0 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 1.3 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.1 | 2.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.4 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 2.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 4.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.4 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.3 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.1 | 0.5 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.8 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.5 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.3 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.1 | 1.7 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 0.2 | GO:0000035 | acyl binding(GO:0000035) |
| 0.1 | 1.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.8 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 0.2 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.1 | 0.7 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 3.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.5 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 1.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 1.3 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.1 | 1.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 0.3 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 0.2 | GO:1904599 | advanced glycation end-product binding(GO:1904599) |
| 0.1 | 1.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 0.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.4 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.1 | 0.5 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.2 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 1.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.7 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 1.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 19.6 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.1 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) |
| 0.0 | 0.3 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 1.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 1.2 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 3.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.7 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.0 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.0 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.0 | 1.7 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 1.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.5 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 1.1 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.2 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.2 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 1.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 1.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 3.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 3.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 2.1 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.5 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 4.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.8 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.3 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 1.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0016279 | protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.4 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 20.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 4.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.1 | 5.0 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 6.1 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 1.0 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 1.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 2.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.8 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 2.3 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 2.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 2.1 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 2.0 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.6 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 2.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.7 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.2 | 6.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 3.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 2.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 4.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 2.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 3.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.3 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 2.8 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 1.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.8 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 2.5 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 1.9 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.0 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 0.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 0.7 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 1.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 2.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 3.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.7 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.4 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.9 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 1.0 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.6 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.0 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.8 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |