Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
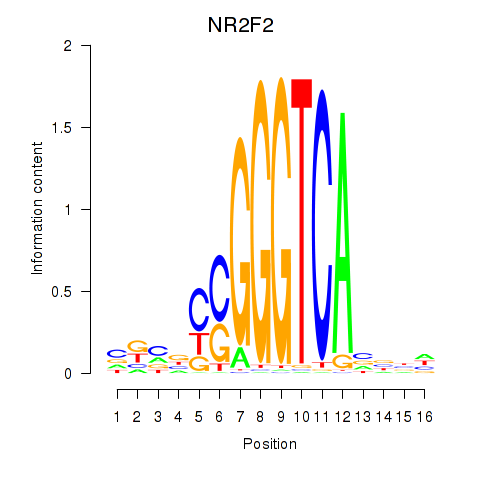
Results for NR2F2
Z-value: 0.62
Transcription factors associated with NR2F2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2F2
|
ENSG00000185551.8 | nuclear receptor subfamily 2 group F member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2F2 | hg19_v2_chr15_+_96876340_96876392 | 0.58 | 2.2e-03 | Click! |
Activity profile of NR2F2 motif
Sorted Z-values of NR2F2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_30960915 | 1.80 |
ENST00000441328.2
ENST00000409899.1 ENST00000409611.1 |
AQP1
|
aquaporin 1 (Colton blood group) |
| chr6_+_142623063 | 1.30 |
ENST00000296932.8
ENST00000367609.3 |
GPR126
|
G protein-coupled receptor 126 |
| chr7_+_79764104 | 1.07 |
ENST00000351004.3
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr20_-_23030296 | 1.07 |
ENST00000377103.2
|
THBD
|
thrombomodulin |
| chr6_+_160183492 | 0.95 |
ENST00000541436.1
|
ACAT2
|
acetyl-CoA acetyltransferase 2 |
| chr7_+_155090271 | 0.91 |
ENST00000476756.1
|
INSIG1
|
insulin induced gene 1 |
| chr13_-_49107303 | 0.89 |
ENST00000344532.3
|
RCBTB2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr1_+_78354297 | 0.83 |
ENST00000334785.7
|
NEXN
|
nexilin (F actin binding protein) |
| chr4_-_186697044 | 0.81 |
ENST00000437304.2
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_+_189157498 | 0.81 |
ENST00000359135.3
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr19_+_41699135 | 0.79 |
ENST00000542619.1
ENST00000600561.1 |
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr4_-_99579733 | 0.77 |
ENST00000305798.3
|
TSPAN5
|
tetraspanin 5 |
| chr1_+_25870070 | 0.75 |
ENST00000374338.4
|
LDLRAP1
|
low density lipoprotein receptor adaptor protein 1 |
| chr4_+_144258021 | 0.74 |
ENST00000262994.4
|
GAB1
|
GRB2-associated binding protein 1 |
| chr6_+_53659746 | 0.69 |
ENST00000370888.1
|
LRRC1
|
leucine rich repeat containing 1 |
| chr17_-_34122596 | 0.68 |
ENST00000250144.8
|
MMP28
|
matrix metallopeptidase 28 |
| chr19_-_18995029 | 0.68 |
ENST00000596048.1
|
CERS1
|
ceramide synthase 1 |
| chr21_+_45553484 | 0.66 |
ENST00000291577.6
ENST00000427803.2 |
C21orf33
|
chromosome 21 open reading frame 33 |
| chr11_-_2906979 | 0.65 |
ENST00000380725.1
ENST00000313407.6 ENST00000430149.2 ENST00000440480.2 ENST00000414822.3 |
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
| chr8_-_124553437 | 0.64 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr17_+_78075361 | 0.64 |
ENST00000577106.1
ENST00000390015.3 |
GAA
|
glucosidase, alpha; acid |
| chr7_+_94139105 | 0.63 |
ENST00000297273.4
|
CASD1
|
CAS1 domain containing 1 |
| chr10_-_131762105 | 0.62 |
ENST00000368648.3
ENST00000355311.5 |
EBF3
|
early B-cell factor 3 |
| chr17_+_78075498 | 0.60 |
ENST00000302262.3
|
GAA
|
glucosidase, alpha; acid |
| chr1_-_236228403 | 0.60 |
ENST00000366595.3
|
NID1
|
nidogen 1 |
| chr5_-_177659539 | 0.59 |
ENST00000476170.2
|
PHYKPL
|
5-phosphohydroxy-L-lysine phospho-lyase |
| chr12_-_110011288 | 0.59 |
ENST00000540016.1
ENST00000266839.5 |
MMAB
|
methylmalonic aciduria (cobalamin deficiency) cblB type |
| chr2_-_264024 | 0.59 |
ENST00000403712.2
ENST00000356150.5 ENST00000405430.1 |
SH3YL1
|
SH3 and SYLF domain containing 1 |
| chr1_-_174992544 | 0.59 |
ENST00000476371.1
|
MRPS14
|
mitochondrial ribosomal protein S14 |
| chr9_+_139971921 | 0.57 |
ENST00000409858.3
|
UAP1L1
|
UDP-N-acteylglucosamine pyrophosphorylase 1-like 1 |
| chr22_+_44319619 | 0.57 |
ENST00000216180.3
|
PNPLA3
|
patatin-like phospholipase domain containing 3 |
| chr1_+_185703513 | 0.56 |
ENST00000271588.4
ENST00000367492.2 |
HMCN1
|
hemicentin 1 |
| chr12_+_56661461 | 0.55 |
ENST00000546544.1
ENST00000553234.1 |
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae) |
| chr8_-_99129338 | 0.55 |
ENST00000520507.1
|
HRSP12
|
heat-responsive protein 12 |
| chr19_-_54693401 | 0.53 |
ENST00000338624.6
|
MBOAT7
|
membrane bound O-acyltransferase domain containing 7 |
| chr11_-_130184470 | 0.52 |
ENST00000357899.4
ENST00000397753.1 |
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr2_+_11052054 | 0.52 |
ENST00000295082.1
|
KCNF1
|
potassium voltage-gated channel, subfamily F, member 1 |
| chr11_-_74022658 | 0.51 |
ENST00000427714.2
ENST00000331597.4 |
P4HA3
|
prolyl 4-hydroxylase, alpha polypeptide III |
| chr22_+_44319648 | 0.51 |
ENST00000423180.2
|
PNPLA3
|
patatin-like phospholipase domain containing 3 |
| chr12_+_56660633 | 0.51 |
ENST00000308197.5
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae) |
| chr9_+_99690592 | 0.51 |
ENST00000354649.3
|
NUTM2G
|
NUT family member 2G |
| chr3_-_122512619 | 0.50 |
ENST00000383659.1
ENST00000306103.2 |
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1 |
| chr19_+_17337406 | 0.50 |
ENST00000597836.1
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr2_+_189157536 | 0.50 |
ENST00000409580.1
ENST00000409637.3 |
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr10_+_94608245 | 0.50 |
ENST00000443748.2
ENST00000260762.6 |
EXOC6
|
exocyst complex component 6 |
| chr12_+_110011571 | 0.50 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr1_-_33168336 | 0.50 |
ENST00000373484.3
|
SYNC
|
syncoilin, intermediate filament protein |
| chr17_-_76124711 | 0.50 |
ENST00000306591.7
ENST00000590602.1 |
TMC6
|
transmembrane channel-like 6 |
| chr7_-_21985656 | 0.50 |
ENST00000406877.3
|
CDCA7L
|
cell division cycle associated 7-like |
| chr22_+_19701985 | 0.49 |
ENST00000455784.2
ENST00000406395.1 |
SEPT5
|
septin 5 |
| chr14_-_75536182 | 0.49 |
ENST00000555463.1
|
ACYP1
|
acylphosphatase 1, erythrocyte (common) type |
| chr13_-_101327028 | 0.49 |
ENST00000328767.5
ENST00000342624.5 ENST00000376234.3 ENST00000423847.1 |
TMTC4
|
transmembrane and tetratricopeptide repeat containing 4 |
| chr19_-_12992244 | 0.49 |
ENST00000538460.1
|
DNASE2
|
deoxyribonuclease II, lysosomal |
| chr12_+_56661033 | 0.49 |
ENST00000433805.2
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae) |
| chr8_-_99129384 | 0.48 |
ENST00000521560.1
ENST00000254878.3 |
HRSP12
|
heat-responsive protein 12 |
| chr17_-_61523535 | 0.48 |
ENST00000584031.1
ENST00000392976.1 |
CYB561
|
cytochrome b561 |
| chr19_-_23869970 | 0.48 |
ENST00000601010.1
|
ZNF675
|
zinc finger protein 675 |
| chr22_+_25465786 | 0.48 |
ENST00000401395.1
|
KIAA1671
|
KIAA1671 |
| chr3_+_45067659 | 0.48 |
ENST00000296130.4
|
CLEC3B
|
C-type lectin domain family 3, member B |
| chr9_-_139581848 | 0.48 |
ENST00000538402.1
ENST00000371694.3 |
AGPAT2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 |
| chr9_-_139581875 | 0.47 |
ENST00000371696.2
|
AGPAT2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 |
| chr7_+_30068260 | 0.47 |
ENST00000440706.2
|
PLEKHA8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| chr4_+_144257915 | 0.47 |
ENST00000262995.4
|
GAB1
|
GRB2-associated binding protein 1 |
| chr19_+_17337027 | 0.46 |
ENST00000601529.1
ENST00000600232.1 |
OCEL1
|
occludin/ELL domain containing 1 |
| chr21_-_47648665 | 0.46 |
ENST00000450351.1
ENST00000522411.1 ENST00000356396.4 ENST00000397728.3 ENST00000457828.2 |
LSS
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) |
| chr21_+_45553535 | 0.45 |
ENST00000348499.5
ENST00000389690.3 ENST00000449622.1 |
C21orf33
|
chromosome 21 open reading frame 33 |
| chrX_+_100333709 | 0.44 |
ENST00000372930.4
|
TMEM35
|
transmembrane protein 35 |
| chr14_-_100841670 | 0.44 |
ENST00000557297.1
ENST00000555813.1 ENST00000557135.1 ENST00000556698.1 ENST00000554509.1 ENST00000555410.1 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr10_+_70320413 | 0.44 |
ENST00000373644.4
|
TET1
|
tet methylcytosine dioxygenase 1 |
| chr19_+_17337007 | 0.43 |
ENST00000215061.4
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr17_-_76124812 | 0.43 |
ENST00000592063.1
ENST00000589271.1 ENST00000322933.4 ENST00000589553.1 |
TMC6
|
transmembrane channel-like 6 |
| chr19_+_10131437 | 0.43 |
ENST00000587782.1
|
RDH8
|
retinol dehydrogenase 8 (all-trans) |
| chr3_-_101232019 | 0.43 |
ENST00000394095.2
ENST00000394091.1 ENST00000394094.2 ENST00000358203.3 ENST00000348610.3 ENST00000314261.7 |
SENP7
|
SUMO1/sentrin specific peptidase 7 |
| chr19_-_35454953 | 0.43 |
ENST00000404801.1
|
ZNF792
|
zinc finger protein 792 |
| chr12_-_125348329 | 0.42 |
ENST00000546215.1
ENST00000415380.2 ENST00000261693.6 ENST00000376788.1 ENST00000545493.1 |
SCARB1
|
scavenger receptor class B, member 1 |
| chr9_-_79009414 | 0.42 |
ENST00000376736.1
|
RFK
|
riboflavin kinase |
| chr8_-_29120580 | 0.41 |
ENST00000524189.1
|
KIF13B
|
kinesin family member 13B |
| chr6_-_2971792 | 0.41 |
ENST00000380546.3
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| chrX_+_54834791 | 0.41 |
ENST00000218439.4
ENST00000375058.1 ENST00000375060.1 |
MAGED2
|
melanoma antigen family D, 2 |
| chr22_-_29137771 | 0.41 |
ENST00000439200.1
ENST00000405598.1 ENST00000398017.2 ENST00000425190.2 ENST00000348295.3 ENST00000382578.1 ENST00000382565.1 ENST00000382566.1 ENST00000382580.2 ENST00000328354.6 |
CHEK2
|
checkpoint kinase 2 |
| chr6_-_2971429 | 0.40 |
ENST00000380529.1
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| chr9_-_79009048 | 0.39 |
ENST00000490113.1
|
RFK
|
riboflavin kinase |
| chr1_-_23694794 | 0.39 |
ENST00000374608.3
|
ZNF436
|
zinc finger protein 436 |
| chr6_+_96025341 | 0.39 |
ENST00000369293.1
ENST00000358812.4 |
MANEA
|
mannosidase, endo-alpha |
| chr17_-_53809473 | 0.39 |
ENST00000575734.1
|
TMEM100
|
transmembrane protein 100 |
| chr5_-_176433693 | 0.39 |
ENST00000507513.1
ENST00000511320.1 |
UIMC1
|
ubiquitin interaction motif containing 1 |
| chr1_-_17307173 | 0.38 |
ENST00000438542.1
ENST00000375535.3 |
MFAP2
|
microfibrillar-associated protein 2 |
| chr7_-_21985489 | 0.38 |
ENST00000356195.5
ENST00000447180.1 ENST00000373934.4 ENST00000457951.1 |
CDCA7L
|
cell division cycle associated 7-like |
| chr17_-_61523622 | 0.38 |
ENST00000448884.2
ENST00000582297.1 ENST00000582034.1 ENST00000578072.1 ENST00000360793.3 |
CYB561
|
cytochrome b561 |
| chr9_+_124030338 | 0.38 |
ENST00000449773.1
ENST00000432226.1 ENST00000436847.1 ENST00000394353.2 ENST00000449733.1 ENST00000412819.1 ENST00000341272.2 ENST00000373808.2 |
GSN
|
gelsolin |
| chr6_-_80657292 | 0.37 |
ENST00000369816.4
|
ELOVL4
|
ELOVL fatty acid elongase 4 |
| chr2_+_85811525 | 0.37 |
ENST00000306384.4
|
VAMP5
|
vesicle-associated membrane protein 5 |
| chr22_-_50700140 | 0.37 |
ENST00000215659.8
|
MAPK12
|
mitogen-activated protein kinase 12 |
| chr15_+_82722225 | 0.37 |
ENST00000300515.8
|
GOLGA6L9
|
golgin A6 family-like 9 |
| chrX_+_54834004 | 0.37 |
ENST00000375068.1
|
MAGED2
|
melanoma antigen family D, 2 |
| chrX_+_54834159 | 0.36 |
ENST00000375053.2
ENST00000347546.4 ENST00000375062.4 |
MAGED2
|
melanoma antigen family D, 2 |
| chr18_+_77160282 | 0.36 |
ENST00000318065.5
ENST00000545796.1 ENST00000592223.1 ENST00000329101.4 ENST00000586434.1 |
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr17_-_18266765 | 0.36 |
ENST00000354098.3
|
SHMT1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr9_+_131038425 | 0.36 |
ENST00000320188.5
ENST00000608796.1 ENST00000419867.2 ENST00000418976.1 |
SWI5
|
SWI5 recombination repair homolog (yeast) |
| chrX_+_152240819 | 0.36 |
ENST00000421798.3
ENST00000535416.1 |
PNMA6C
PNMA6A
|
paraneoplastic Ma antigen family member 6C paraneoplastic Ma antigen family member 6A |
| chr4_-_107237374 | 0.36 |
ENST00000361687.4
ENST00000507696.1 ENST00000394708.2 ENST00000509532.1 |
TBCK
|
TBC1 domain containing kinase |
| chr3_-_142607740 | 0.36 |
ENST00000485766.1
|
PCOLCE2
|
procollagen C-endopeptidase enhancer 2 |
| chr22_+_19705928 | 0.36 |
ENST00000383045.3
ENST00000438754.2 |
SEPT5
|
septin 5 |
| chr15_+_83098710 | 0.35 |
ENST00000561062.1
ENST00000358583.3 |
GOLGA6L9
|
golgin A6 family-like 20 |
| chr4_-_107237340 | 0.35 |
ENST00000394706.3
|
TBCK
|
TBC1 domain containing kinase |
| chr6_-_2971494 | 0.35 |
ENST00000380539.1
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| chr22_-_37584321 | 0.34 |
ENST00000397110.2
ENST00000337843.2 |
C1QTNF6
|
C1q and tumor necrosis factor related protein 6 |
| chr12_+_51632666 | 0.34 |
ENST00000604900.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr4_-_18023350 | 0.34 |
ENST00000539056.1
ENST00000382226.5 ENST00000326877.4 |
LCORL
|
ligand dependent nuclear receptor corepressor-like |
| chrX_+_54835493 | 0.34 |
ENST00000396224.1
|
MAGED2
|
melanoma antigen family D, 2 |
| chr18_-_268019 | 0.33 |
ENST00000261600.6
|
THOC1
|
THO complex 1 |
| chr1_+_215740709 | 0.33 |
ENST00000259154.4
|
KCTD3
|
potassium channel tetramerization domain containing 3 |
| chr3_-_50340996 | 0.33 |
ENST00000266031.4
ENST00000395143.2 ENST00000457214.2 ENST00000447605.2 ENST00000418723.1 ENST00000395144.2 |
HYAL1
|
hyaluronoglucosaminidase 1 |
| chr22_-_19165917 | 0.32 |
ENST00000451283.1
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 |
| chrX_-_107018969 | 0.32 |
ENST00000372383.4
|
TSC22D3
|
TSC22 domain family, member 3 |
| chr2_+_61108650 | 0.32 |
ENST00000295025.8
|
REL
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr14_+_24702099 | 0.32 |
ENST00000420554.2
|
GMPR2
|
guanosine monophosphate reductase 2 |
| chr17_-_61777090 | 0.32 |
ENST00000578061.1
|
LIMD2
|
LIM domain containing 2 |
| chr1_+_207925391 | 0.32 |
ENST00000358170.2
ENST00000354848.1 ENST00000322918.5 ENST00000367042.1 ENST00000367041.1 ENST00000357714.1 ENST00000322875.4 ENST00000367047.1 ENST00000441839.2 ENST00000361067.1 ENST00000360212.2 ENST00000480003.1 |
CD46
|
CD46 molecule, complement regulatory protein |
| chr10_-_97321112 | 0.32 |
ENST00000607232.1
ENST00000371227.4 ENST00000371249.2 ENST00000371247.2 ENST00000371246.2 ENST00000393949.1 ENST00000353505.5 ENST00000347291.4 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr19_+_19639670 | 0.31 |
ENST00000436027.5
|
YJEFN3
|
YjeF N-terminal domain containing 3 |
| chr19_-_49926698 | 0.31 |
ENST00000270631.1
|
PTH2
|
parathyroid hormone 2 |
| chr1_-_151882031 | 0.31 |
ENST00000489410.1
|
THEM4
|
thioesterase superfamily member 4 |
| chr17_-_18266660 | 0.31 |
ENST00000582653.1
ENST00000352886.6 |
SHMT1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr2_+_61108771 | 0.30 |
ENST00000394479.3
|
REL
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr19_+_18118972 | 0.30 |
ENST00000593560.2
ENST00000222250.4 |
ARRDC2
|
arrestin domain containing 2 |
| chr14_-_103987679 | 0.30 |
ENST00000553610.1
|
CKB
|
creatine kinase, brain |
| chr10_-_97321165 | 0.30 |
ENST00000306402.6
|
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr9_-_86432547 | 0.30 |
ENST00000376365.3
ENST00000376371.2 |
GKAP1
|
G kinase anchoring protein 1 |
| chr17_-_5026397 | 0.30 |
ENST00000250076.3
|
ZNF232
|
zinc finger protein 232 |
| chr11_-_65308082 | 0.30 |
ENST00000532661.1
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr1_+_33207381 | 0.29 |
ENST00000401073.2
|
KIAA1522
|
KIAA1522 |
| chr19_-_2050852 | 0.29 |
ENST00000541165.1
ENST00000591601.1 |
MKNK2
|
MAP kinase interacting serine/threonine kinase 2 |
| chr9_-_33264676 | 0.29 |
ENST00000472232.3
ENST00000379704.2 |
BAG1
|
BCL2-associated athanogene |
| chr1_-_15850676 | 0.29 |
ENST00000440484.1
ENST00000333868.5 |
CASP9
|
caspase 9, apoptosis-related cysteine peptidase |
| chr11_-_65325430 | 0.29 |
ENST00000322147.4
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr5_-_133747589 | 0.28 |
ENST00000458198.2
|
CDKN2AIPNL
|
CDKN2A interacting protein N-terminal like |
| chr12_-_108954933 | 0.28 |
ENST00000431469.2
ENST00000546815.1 |
SART3
|
squamous cell carcinoma antigen recognized by T cells 3 |
| chr16_+_66613351 | 0.28 |
ENST00000379486.2
ENST00000268595.2 |
CMTM2
|
CKLF-like MARVEL transmembrane domain containing 2 |
| chr9_-_116840728 | 0.28 |
ENST00000265132.3
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr9_-_125027079 | 0.28 |
ENST00000417201.3
|
RBM18
|
RNA binding motif protein 18 |
| chr16_+_524850 | 0.28 |
ENST00000450428.1
ENST00000452814.1 |
RAB11FIP3
|
RAB11 family interacting protein 3 (class II) |
| chr19_-_12992274 | 0.28 |
ENST00000592506.1
ENST00000222219.3 |
DNASE2
|
deoxyribonuclease II, lysosomal |
| chr13_+_103451399 | 0.28 |
ENST00000257336.1
ENST00000448849.2 |
BIVM
|
basic, immunoglobulin-like variable motif containing |
| chr16_-_89785777 | 0.27 |
ENST00000561976.1
|
VPS9D1
|
VPS9 domain containing 1 |
| chr15_-_66790146 | 0.27 |
ENST00000316634.5
|
SNAPC5
|
small nuclear RNA activating complex, polypeptide 5, 19kDa |
| chr11_-_27722021 | 0.27 |
ENST00000356660.4
ENST00000418212.1 ENST00000533246.1 |
BDNF
|
brain-derived neurotrophic factor |
| chr15_-_37393406 | 0.27 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr16_+_57702210 | 0.27 |
ENST00000450388.3
|
GPR97
|
G protein-coupled receptor 97 |
| chr1_+_155036204 | 0.27 |
ENST00000368409.3
ENST00000359751.4 ENST00000427683.2 ENST00000556931.1 ENST00000505139.1 |
EFNA4
EFNA3
EFNA3
|
ephrin-A4 ephrin-A3 Ephrin-A3; Uncharacterized protein; cDNA FLJ57652, highly similar to Ephrin-A3 |
| chr7_+_56131917 | 0.27 |
ENST00000434526.2
ENST00000275607.9 ENST00000395435.2 ENST00000413952.2 ENST00000342190.6 ENST00000437307.2 ENST00000413756.1 ENST00000451338.1 |
SUMF2
|
sulfatase modifying factor 2 |
| chr17_+_15902694 | 0.27 |
ENST00000261647.5
ENST00000486880.2 |
TTC19
|
tetratricopeptide repeat domain 19 |
| chr17_-_76356148 | 0.27 |
ENST00000587578.1
ENST00000330871.2 |
SOCS3
|
suppressor of cytokine signaling 3 |
| chr19_-_7040190 | 0.27 |
ENST00000381394.4
|
MBD3L4
|
methyl-CpG binding domain protein 3-like 4 |
| chr19_+_46001697 | 0.26 |
ENST00000451287.2
ENST00000324688.4 |
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr17_+_79373540 | 0.26 |
ENST00000307745.7
|
RP11-1055B8.7
|
BAH and coiled-coil domain-containing protein 1 |
| chr19_-_40030861 | 0.26 |
ENST00000390658.2
|
EID2
|
EP300 interacting inhibitor of differentiation 2 |
| chr19_-_4517613 | 0.26 |
ENST00000301286.3
|
PLIN4
|
perilipin 4 |
| chr3_+_52489503 | 0.26 |
ENST00000345716.4
|
NISCH
|
nischarin |
| chr22_-_29138386 | 0.26 |
ENST00000544772.1
|
CHEK2
|
checkpoint kinase 2 |
| chr7_+_128828713 | 0.26 |
ENST00000249373.3
|
SMO
|
smoothened, frizzled family receptor |
| chr20_-_52687059 | 0.26 |
ENST00000371435.2
ENST00000395961.3 |
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr20_-_23402028 | 0.26 |
ENST00000398425.3
ENST00000432543.2 ENST00000377026.4 |
NAPB
|
N-ethylmaleimide-sensitive factor attachment protein, beta |
| chr15_-_75248954 | 0.25 |
ENST00000499788.2
|
RPP25
|
ribonuclease P/MRP 25kDa subunit |
| chr1_-_3816779 | 0.25 |
ENST00000361605.3
|
C1orf174
|
chromosome 1 open reading frame 174 |
| chr14_+_61447832 | 0.25 |
ENST00000354886.2
ENST00000267488.4 |
SLC38A6
|
solute carrier family 38, member 6 |
| chr18_-_53255766 | 0.25 |
ENST00000566286.1
ENST00000564999.1 ENST00000566279.1 ENST00000354452.3 ENST00000356073.4 |
TCF4
|
transcription factor 4 |
| chr15_-_52263937 | 0.25 |
ENST00000315141.5
ENST00000299601.5 |
LEO1
|
Leo1, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) |
| chr6_-_31865452 | 0.25 |
ENST00000375530.4
ENST00000375537.4 |
EHMT2
|
euchromatic histone-lysine N-methyltransferase 2 |
| chr1_+_179851823 | 0.25 |
ENST00000435319.4
|
TOR1AIP1
|
torsin A interacting protein 1 |
| chr16_+_69458537 | 0.25 |
ENST00000515314.1
ENST00000561792.1 ENST00000568237.1 |
CYB5B
|
cytochrome b5 type B (outer mitochondrial membrane) |
| chr9_+_131037623 | 0.25 |
ENST00000495313.1
ENST00000372898.2 |
SWI5
|
SWI5 recombination repair homolog (yeast) |
| chr8_-_77912431 | 0.25 |
ENST00000357039.4
ENST00000522527.1 |
PEX2
|
peroxisomal biogenesis factor 2 |
| chr13_+_115047097 | 0.24 |
ENST00000351487.5
|
UPF3A
|
UPF3 regulator of nonsense transcripts homolog A (yeast) |
| chr11_-_130184555 | 0.24 |
ENST00000525842.1
|
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr10_+_23728198 | 0.24 |
ENST00000376495.3
|
OTUD1
|
OTU domain containing 1 |
| chr1_-_20812690 | 0.24 |
ENST00000375078.3
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| chr16_+_75182376 | 0.24 |
ENST00000570010.1
ENST00000568079.1 ENST00000464850.1 ENST00000332307.4 ENST00000393430.2 |
ZFP1
|
ZFP1 zinc finger protein |
| chr1_+_76190357 | 0.24 |
ENST00000370834.5
ENST00000541113.1 ENST00000543667.1 ENST00000420607.2 |
ACADM
|
acyl-CoA dehydrogenase, C-4 to C-12 straight chain |
| chr17_-_18266797 | 0.24 |
ENST00000316694.3
ENST00000539052.1 |
SHMT1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr1_-_15850839 | 0.24 |
ENST00000348549.5
ENST00000546424.1 |
CASP9
|
caspase 9, apoptosis-related cysteine peptidase |
| chr16_+_69458428 | 0.23 |
ENST00000512062.1
ENST00000307892.8 |
CYB5B
|
cytochrome b5 type B (outer mitochondrial membrane) |
| chr1_+_10092890 | 0.23 |
ENST00000253251.8
ENST00000377157.3 |
UBE4B
|
ubiquitination factor E4B |
| chr19_+_46000479 | 0.23 |
ENST00000456399.2
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr20_-_3154162 | 0.23 |
ENST00000360342.3
|
LZTS3
|
Homo sapiens leucine zipper, putative tumor suppressor family member 3 (LZTS3), mRNA. |
| chr9_-_135230336 | 0.23 |
ENST00000224140.5
ENST00000372169.2 ENST00000393220.1 |
SETX
|
senataxin |
| chr9_+_96051469 | 0.23 |
ENST00000453718.1
|
WNK2
|
WNK lysine deficient protein kinase 2 |
| chr13_+_115047053 | 0.23 |
ENST00000375299.3
|
UPF3A
|
UPF3 regulator of nonsense transcripts homolog A (yeast) |
| chr20_-_2821271 | 0.23 |
ENST00000448755.1
ENST00000360652.2 |
PCED1A
|
PC-esterase domain containing 1A |
| chr13_+_114462193 | 0.22 |
ENST00000375353.3
|
TMEM255B
|
transmembrane protein 255B |
| chr14_+_64970662 | 0.22 |
ENST00000556965.1
ENST00000554015.1 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr11_-_89224139 | 0.22 |
ENST00000413594.2
|
NOX4
|
NADPH oxidase 4 |
| chr11_-_19082216 | 0.22 |
ENST00000329773.2
|
MRGPRX2
|
MAS-related GPR, member X2 |
| chr1_+_203764742 | 0.22 |
ENST00000432282.1
ENST00000453771.1 ENST00000367214.1 ENST00000367212.3 ENST00000332127.4 |
ZC3H11A
|
zinc finger CCCH-type containing 11A |
| chr6_-_10419871 | 0.22 |
ENST00000319516.4
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr7_+_99746514 | 0.22 |
ENST00000341942.5
ENST00000441173.1 |
LAMTOR4
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 |
| chr8_+_142138799 | 0.21 |
ENST00000518668.1
|
DENND3
|
DENN/MADD domain containing 3 |
| chr1_-_155880672 | 0.21 |
ENST00000609492.1
ENST00000368322.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr10_+_120967072 | 0.21 |
ENST00000392870.2
|
GRK5
|
G protein-coupled receptor kinase 5 |
| chr7_+_36192855 | 0.21 |
ENST00000534978.1
|
EEPD1
|
endonuclease/exonuclease/phosphatase family domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2F2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0072229 | carbon dioxide transmembrane transport(GO:0035378) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.3 | 0.9 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.3 | 0.9 | GO:1904482 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.3 | 1.2 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.3 | 0.8 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) |
| 0.2 | 0.7 | GO:0072720 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.2 | 0.7 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.2 | 0.8 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.2 | 0.5 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.2 | 1.5 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.2 | 0.5 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.2 | 0.5 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 0.6 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.6 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 0.4 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 0.4 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.1 | 0.7 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 0.1 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.1 | 0.3 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.1 | 0.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 0.8 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.1 | 0.3 | GO:0015728 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.1 | 0.3 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.1 | 1.1 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.1 | 0.4 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.3 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 1.3 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.3 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.1 | 0.2 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 1.0 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.1 | 0.4 | GO:0044858 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.1 | 0.3 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 0.2 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.6 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 0.5 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 0.3 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.1 | 0.6 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.3 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.3 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.1 | 0.3 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 0.4 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 0.2 | GO:0015993 | molecular hydrogen transport(GO:0015993) |
| 0.1 | 0.6 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.2 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.1 | 0.2 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.1 | 0.2 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.1 | 0.2 | GO:0030264 | nuclear fragmentation involved in apoptotic nuclear change(GO:0030264) |
| 0.1 | 0.2 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 0.3 | GO:0072093 | positive regulation of hh target transcription factor activity(GO:0007228) ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) metanephric renal vesicle formation(GO:0072093) |
| 0.0 | 0.2 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.2 | GO:0009440 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.0 | 1.1 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.0 | 0.2 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.6 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.8 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.4 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.4 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.2 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.4 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.7 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.6 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.7 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.3 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.4 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.0 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.7 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.6 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.4 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.5 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.2 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.8 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.3 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.2 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.1 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.3 | GO:0032049 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.6 | GO:0006554 | lysine catabolic process(GO:0006554) |
| 0.0 | 0.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:1902994 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.0 | 0.7 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.3 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:1901253 | mesodermal cell fate determination(GO:0007500) negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.0 | 0.3 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.1 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.4 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.8 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.1 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 0.6 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.1 | GO:0061107 | prostate gland stromal morphogenesis(GO:0060741) seminal vesicle development(GO:0061107) |
| 0.0 | 0.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.3 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.8 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0051944 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 0.8 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.1 | GO:0097187 | dentinogenesis(GO:0097187) |
| 0.0 | 0.1 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.3 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.2 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:1904199 | positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.0 | 0.3 | GO:0035581 | sequestering of extracellular ligand from receptor(GO:0035581) |
| 0.0 | 0.1 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.0 | 0.2 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.5 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:0050894 | determination of affect(GO:0050894) |
| 0.0 | 0.3 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.2 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.3 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.1 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.1 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 1.0 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 1.7 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 0.1 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 0.1 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.2 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.3 | GO:0048243 | norepinephrine secretion(GO:0048243) |
| 0.0 | 0.8 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.1 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.1 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.1 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.8 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.0 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.3 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.1 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.5 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.1 | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.0 | 0.3 | GO:0031114 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.2 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.3 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:0003222 | ventricular trabecula myocardium morphogenesis(GO:0003222) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.1 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) |
| 0.0 | 0.0 | GO:0014063 | regulation of serotonin secretion(GO:0014062) negative regulation of serotonin secretion(GO:0014063) |
| 0.0 | 0.1 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.2 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.1 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.5 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:1902957 | negative regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902957) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.2 | 0.9 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 0.6 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.1 | 0.5 | GO:0097196 | Shu complex(GO:0097196) |
| 0.1 | 0.5 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 0.9 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.6 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.4 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 0.3 | GO:0071745 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 0.1 | 0.5 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 0.3 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 0.3 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 0.2 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.1 | 0.3 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.3 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.0 | 0.2 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.4 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 1.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 1.6 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.6 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 2.9 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.3 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.5 | GO:0043186 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.7 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.6 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.0 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 1.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.6 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 2.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 1.0 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.3 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.7 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.4 | 1.2 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.3 | 0.8 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.2 | 1.0 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.2 | 0.9 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.2 | 0.9 | GO:0070905 | serine binding(GO:0070905) |
| 0.2 | 1.5 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.2 | 0.8 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 0.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.4 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 1.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 1.0 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 0.6 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.1 | 0.5 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.1 | 0.3 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.1 | 0.3 | GO:0052857 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.1 | 0.3 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.1 | 0.3 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 1.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.4 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 0.3 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.5 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.2 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.1 | 0.2 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 0.2 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 0.3 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.6 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.1 | 0.4 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.2 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.1 | 1.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.2 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.1 | 0.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.2 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.1 | 0.2 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 0.8 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 0.3 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 0.3 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.2 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.1 | 0.6 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.0 | 0.4 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.4 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.1 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.3 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0001003 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.1 | GO:0052825 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.0 | 0.3 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0090556 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.4 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.3 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.1 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.1 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) |
| 0.0 | 0.1 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.3 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.0 | 0.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.0 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.7 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.9 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.3 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.5 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.0 | 0.4 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.0 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.5 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 1.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.9 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.5 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.8 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.4 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.8 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | PID FOXO PATHWAY | FoxO family signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.7 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 1.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.7 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.9 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |