Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
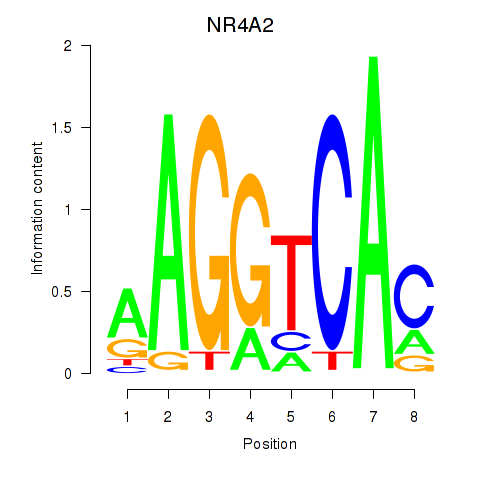
Results for NR4A2
Z-value: 0.56
Transcription factors associated with NR4A2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR4A2
|
ENSG00000153234.9 | nuclear receptor subfamily 4 group A member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR4A2 | hg19_v2_chr2_-_157189180_157189290 | 0.05 | 8.1e-01 | Click! |
Activity profile of NR4A2 motif
Sorted Z-values of NR4A2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_228678550 | 3.11 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr10_+_81107271 | 1.08 |
ENST00000448165.1
|
PPIF
|
peptidylprolyl isomerase F |
| chr14_-_55369525 | 1.03 |
ENST00000543643.2
ENST00000536224.2 ENST00000395514.1 ENST00000491895.2 |
GCH1
|
GTP cyclohydrolase 1 |
| chr1_+_212738676 | 0.91 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr19_+_45251804 | 0.88 |
ENST00000164227.5
|
BCL3
|
B-cell CLL/lymphoma 3 |
| chr13_+_97928395 | 0.85 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr2_+_162016827 | 0.78 |
ENST00000429217.1
ENST00000406287.1 ENST00000402568.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr4_-_139163491 | 0.71 |
ENST00000280612.5
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr9_-_136344237 | 0.69 |
ENST00000432868.1
ENST00000371899.4 |
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr12_-_120554534 | 0.64 |
ENST00000538903.1
ENST00000534951.1 |
RAB35
|
RAB35, member RAS oncogene family |
| chr15_-_55562582 | 0.61 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr2_+_11817713 | 0.58 |
ENST00000449576.2
|
LPIN1
|
lipin 1 |
| chr2_+_162016804 | 0.57 |
ENST00000392749.2
ENST00000440506.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr1_+_169075554 | 0.57 |
ENST00000367815.4
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr15_-_55562479 | 0.54 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr9_-_136344197 | 0.52 |
ENST00000414172.1
ENST00000371897.4 |
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr7_+_112090483 | 0.50 |
ENST00000403825.3
ENST00000429071.1 |
IFRD1
|
interferon-related developmental regulator 1 |
| chr2_+_162016916 | 0.47 |
ENST00000405852.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr20_+_37590942 | 0.46 |
ENST00000373325.2
ENST00000252011.3 ENST00000373323.4 |
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr5_-_131347501 | 0.45 |
ENST00000543479.1
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr11_-_47470591 | 0.45 |
ENST00000524487.1
|
RAPSN
|
receptor-associated protein of the synapse |
| chr8_+_21911054 | 0.45 |
ENST00000519850.1
ENST00000381470.3 |
DMTN
|
dematin actin binding protein |
| chr7_+_128399002 | 0.44 |
ENST00000493278.1
|
CALU
|
calumenin |
| chr8_+_145149930 | 0.43 |
ENST00000318911.4
|
CYC1
|
cytochrome c-1 |
| chr10_+_81107216 | 0.42 |
ENST00000394579.3
ENST00000225174.3 |
PPIF
|
peptidylprolyl isomerase F |
| chr12_-_46121554 | 0.41 |
ENST00000609803.1
|
LINC00938
|
long intergenic non-protein coding RNA 938 |
| chr13_+_102142296 | 0.41 |
ENST00000376162.3
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr19_+_1407733 | 0.38 |
ENST00000592453.1
|
DAZAP1
|
DAZ associated protein 1 |
| chr3_-_113464906 | 0.37 |
ENST00000477813.1
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr6_-_34664612 | 0.37 |
ENST00000374023.3
ENST00000374026.3 |
C6orf106
|
chromosome 6 open reading frame 106 |
| chr22_+_30792980 | 0.37 |
ENST00000403484.1
ENST00000405717.3 ENST00000402592.3 |
SEC14L2
|
SEC14-like 2 (S. cerevisiae) |
| chr1_-_27816556 | 0.36 |
ENST00000536657.1
|
WASF2
|
WAS protein family, member 2 |
| chr1_-_157108266 | 0.35 |
ENST00000326786.4
|
ETV3
|
ets variant 3 |
| chr8_+_134125727 | 0.35 |
ENST00000521107.1
|
TG
|
thyroglobulin |
| chr3_+_113465866 | 0.35 |
ENST00000273398.3
ENST00000538620.1 ENST00000496747.1 ENST00000475322.1 |
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
| chr5_+_66124590 | 0.35 |
ENST00000490016.2
ENST00000403666.1 ENST00000450827.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr11_+_67007518 | 0.34 |
ENST00000530342.1
ENST00000308783.5 |
KDM2A
|
lysine (K)-specific demethylase 2A |
| chr1_+_206858328 | 0.34 |
ENST00000367103.3
|
MAPKAPK2
|
mitogen-activated protein kinase-activated protein kinase 2 |
| chr15_+_59730348 | 0.33 |
ENST00000288228.5
ENST00000559628.1 ENST00000557914.1 ENST00000560474.1 |
FAM81A
|
family with sequence similarity 81, member A |
| chrX_-_150067069 | 0.32 |
ENST00000466436.1
|
CD99L2
|
CD99 molecule-like 2 |
| chr11_-_47470703 | 0.31 |
ENST00000298854.2
|
RAPSN
|
receptor-associated protein of the synapse |
| chr1_+_206858232 | 0.30 |
ENST00000294981.4
|
MAPKAPK2
|
mitogen-activated protein kinase-activated protein kinase 2 |
| chr16_+_85942594 | 0.30 |
ENST00000566369.1
|
IRF8
|
interferon regulatory factor 8 |
| chr17_+_7533439 | 0.30 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr12_+_120875910 | 0.30 |
ENST00000551806.1
|
AL021546.6
|
Glutamyl-tRNA(Gln) amidotransferase subunit C, mitochondrial |
| chr12_-_81992111 | 0.29 |
ENST00000443686.3
ENST00000407050.4 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr14_+_23791159 | 0.28 |
ENST00000557702.1
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr20_+_43343886 | 0.27 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr14_+_21498666 | 0.27 |
ENST00000481535.1
|
TPPP2
|
tubulin polymerization-promoting protein family member 2 |
| chr20_-_48532019 | 0.27 |
ENST00000289431.5
|
SPATA2
|
spermatogenesis associated 2 |
| chr6_-_39290316 | 0.27 |
ENST00000425054.2
ENST00000373227.4 ENST00000373229.5 ENST00000437525.2 |
KCNK16
|
potassium channel, subfamily K, member 16 |
| chr19_+_6739662 | 0.24 |
ENST00000313285.8
ENST00000313244.9 ENST00000596758.1 |
TRIP10
|
thyroid hormone receptor interactor 10 |
| chr10_+_51371390 | 0.24 |
ENST00000478381.1
ENST00000451577.2 ENST00000374098.2 ENST00000374097.2 |
TIMM23B
|
translocase of inner mitochondrial membrane 23 homolog B (yeast) |
| chr11_-_47470682 | 0.24 |
ENST00000529341.1
ENST00000352508.3 |
RAPSN
|
receptor-associated protein of the synapse |
| chr17_-_27503770 | 0.24 |
ENST00000533112.1
|
MYO18A
|
myosin XVIIIA |
| chr12_-_108954933 | 0.24 |
ENST00000431469.2
ENST00000546815.1 |
SART3
|
squamous cell carcinoma antigen recognized by T cells 3 |
| chr22_+_30821732 | 0.23 |
ENST00000355143.4
|
MTFP1
|
mitochondrial fission process 1 |
| chr10_-_50970322 | 0.23 |
ENST00000374103.4
|
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr3_+_129247479 | 0.23 |
ENST00000296271.3
|
RHO
|
rhodopsin |
| chr15_-_41408339 | 0.23 |
ENST00000401393.3
|
INO80
|
INO80 complex subunit |
| chr19_+_46850251 | 0.23 |
ENST00000012443.4
|
PPP5C
|
protein phosphatase 5, catalytic subunit |
| chr19_+_45394477 | 0.23 |
ENST00000252487.5
ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr10_-_50970382 | 0.23 |
ENST00000419399.1
ENST00000432695.1 |
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr20_+_31755934 | 0.22 |
ENST00000354932.5
|
BPIFA2
|
BPI fold containing family A, member 2 |
| chr16_+_4674787 | 0.22 |
ENST00000262370.7
|
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr5_-_131347583 | 0.22 |
ENST00000379255.1
ENST00000430403.1 ENST00000544770.1 ENST00000379246.1 ENST00000414078.1 ENST00000441995.1 |
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr17_+_73975292 | 0.22 |
ENST00000397640.1
ENST00000416485.1 ENST00000588202.1 ENST00000590676.1 ENST00000586891.1 |
TEN1
|
TEN1 CST complex subunit |
| chr19_-_4535233 | 0.21 |
ENST00000381848.3
ENST00000588887.1 ENST00000586133.1 |
PLIN5
|
perilipin 5 |
| chr1_+_155583012 | 0.21 |
ENST00000462250.2
|
MSTO1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr1_-_1711508 | 0.21 |
ENST00000378625.1
|
NADK
|
NAD kinase |
| chr22_+_18121356 | 0.20 |
ENST00000317582.5
ENST00000543133.1 ENST00000538149.1 ENST00000337612.5 ENST00000493680.1 |
BCL2L13
|
BCL2-like 13 (apoptosis facilitator) |
| chr19_+_42824511 | 0.20 |
ENST00000601644.1
|
TMEM145
|
transmembrane protein 145 |
| chr1_-_8877692 | 0.20 |
ENST00000400908.2
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats |
| chr9_-_39239171 | 0.20 |
ENST00000358144.2
|
CNTNAP3
|
contactin associated protein-like 3 |
| chr22_-_39239987 | 0.20 |
ENST00000333039.2
|
NPTXR
|
neuronal pentraxin receptor |
| chr3_+_14989186 | 0.19 |
ENST00000435454.1
ENST00000323373.6 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr19_+_3880581 | 0.19 |
ENST00000450849.2
ENST00000301260.6 ENST00000398448.3 |
ATCAY
|
ataxia, cerebellar, Cayman type |
| chrX_+_135230712 | 0.19 |
ENST00000535737.1
|
FHL1
|
four and a half LIM domains 1 |
| chr3_-_197024965 | 0.19 |
ENST00000392382.2
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr2_+_220299547 | 0.19 |
ENST00000312358.7
|
SPEG
|
SPEG complex locus |
| chr9_-_130487143 | 0.19 |
ENST00000419060.1
|
PTRH1
|
peptidyl-tRNA hydrolase 1 homolog (S. cerevisiae) |
| chr2_-_26467465 | 0.19 |
ENST00000457468.2
|
HADHA
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit |
| chr22_+_50354104 | 0.19 |
ENST00000360612.4
|
PIM3
|
pim-3 oncogene |
| chr11_+_67798363 | 0.18 |
ENST00000525628.1
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr7_+_26591441 | 0.18 |
ENST00000420912.1
ENST00000457000.1 ENST00000430426.1 |
AC004947.2
|
AC004947.2 |
| chr1_-_229569834 | 0.18 |
ENST00000366684.3
ENST00000366683.2 |
ACTA1
|
actin, alpha 1, skeletal muscle |
| chr22_-_36902522 | 0.18 |
ENST00000397223.4
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr17_-_49021974 | 0.17 |
ENST00000501718.2
|
RP11-700H6.1
|
RP11-700H6.1 |
| chr8_+_22429205 | 0.17 |
ENST00000520207.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr1_+_36690011 | 0.16 |
ENST00000354618.5
ENST00000469141.2 ENST00000478853.1 |
THRAP3
|
thyroid hormone receptor associated protein 3 |
| chr12_+_51818749 | 0.16 |
ENST00000514353.3
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr17_-_10276319 | 0.16 |
ENST00000252172.4
ENST00000418404.3 |
MYH13
|
myosin, heavy chain 13, skeletal muscle |
| chr9_-_96215822 | 0.16 |
ENST00000375412.5
|
FAM120AOS
|
family with sequence similarity 120A opposite strand |
| chr8_-_145018905 | 0.16 |
ENST00000398774.2
|
PLEC
|
plectin |
| chr17_-_1553346 | 0.16 |
ENST00000301336.6
|
RILP
|
Rab interacting lysosomal protein |
| chr4_+_41614720 | 0.15 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr16_-_70729496 | 0.15 |
ENST00000567648.1
|
VAC14
|
Vac14 homolog (S. cerevisiae) |
| chr11_+_76571911 | 0.15 |
ENST00000534206.1
ENST00000532485.1 ENST00000526597.1 ENST00000533873.1 ENST00000538157.1 |
ACER3
|
alkaline ceramidase 3 |
| chr7_+_73245193 | 0.15 |
ENST00000340958.2
|
CLDN4
|
claudin 4 |
| chr12_-_120554622 | 0.15 |
ENST00000229340.5
|
RAB35
|
RAB35, member RAS oncogene family |
| chr17_+_45973516 | 0.14 |
ENST00000376741.4
|
SP2
|
Sp2 transcription factor |
| chr1_-_157108130 | 0.14 |
ENST00000368192.4
|
ETV3
|
ets variant 3 |
| chr22_-_30960876 | 0.14 |
ENST00000401975.1
ENST00000428682.1 ENST00000423299.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr2_-_220110111 | 0.14 |
ENST00000428427.1
ENST00000356283.3 ENST00000432839.1 ENST00000424620.1 |
GLB1L
|
galactosidase, beta 1-like |
| chr12_+_51818555 | 0.14 |
ENST00000453097.2
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr17_+_36858694 | 0.14 |
ENST00000563897.1
|
CTB-58E17.1
|
CTB-58E17.1 |
| chr3_-_167452262 | 0.14 |
ENST00000487947.2
|
PDCD10
|
programmed cell death 10 |
| chr2_-_207024134 | 0.14 |
ENST00000457011.1
ENST00000440274.1 ENST00000432169.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr4_+_48343339 | 0.14 |
ENST00000264313.6
|
SLAIN2
|
SLAIN motif family, member 2 |
| chr1_-_27816641 | 0.14 |
ENST00000430629.2
|
WASF2
|
WAS protein family, member 2 |
| chr11_-_64511789 | 0.13 |
ENST00000419843.1
ENST00000394430.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr22_+_20104947 | 0.13 |
ENST00000402752.1
|
RANBP1
|
RAN binding protein 1 |
| chr12_+_51818586 | 0.13 |
ENST00000394856.1
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr19_-_55874611 | 0.13 |
ENST00000424985.3
|
FAM71E2
|
family with sequence similarity 71, member E2 |
| chr19_+_4153598 | 0.13 |
ENST00000078445.2
ENST00000252587.3 ENST00000595923.1 ENST00000602257.1 ENST00000602147.1 |
CREB3L3
|
cAMP responsive element binding protein 3-like 3 |
| chr22_-_37880543 | 0.13 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr16_-_85833109 | 0.13 |
ENST00000253457.3
|
EMC8
|
ER membrane protein complex subunit 8 |
| chr6_-_31628512 | 0.13 |
ENST00000375911.1
|
C6orf47
|
chromosome 6 open reading frame 47 |
| chr2_-_207023918 | 0.13 |
ENST00000455934.2
ENST00000449699.1 ENST00000454195.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr15_-_41408409 | 0.12 |
ENST00000361937.3
|
INO80
|
INO80 complex subunit |
| chr3_-_113465065 | 0.12 |
ENST00000497255.1
ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr20_+_57875658 | 0.12 |
ENST00000371025.3
|
EDN3
|
endothelin 3 |
| chr2_-_25194476 | 0.12 |
ENST00000534855.1
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr1_+_201924619 | 0.12 |
ENST00000367287.4
|
TIMM17A
|
translocase of inner mitochondrial membrane 17 homolog A (yeast) |
| chr9_+_95997205 | 0.12 |
ENST00000411624.1
|
WNK2
|
WNK lysine deficient protein kinase 2 |
| chr11_-_78052923 | 0.11 |
ENST00000340149.2
|
GAB2
|
GRB2-associated binding protein 2 |
| chr9_-_34048873 | 0.11 |
ENST00000449054.1
ENST00000379239.4 ENST00000539807.1 ENST00000379238.1 ENST00000418786.2 ENST00000360802.1 ENST00000412543.1 |
UBAP2
|
ubiquitin associated protein 2 |
| chr1_+_26348259 | 0.11 |
ENST00000374280.3
|
EXTL1
|
exostosin-like glycosyltransferase 1 |
| chr15_-_41120896 | 0.11 |
ENST00000299174.5
ENST00000427255.2 |
PPP1R14D
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr8_+_134203303 | 0.11 |
ENST00000519433.1
ENST00000517423.1 ENST00000377863.2 ENST00000220856.6 |
WISP1
|
WNT1 inducible signaling pathway protein 1 |
| chr16_-_66952742 | 0.11 |
ENST00000565235.2
ENST00000568632.1 ENST00000565796.1 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr8_+_145582633 | 0.11 |
ENST00000540505.1
|
SLC52A2
|
solute carrier family 52 (riboflavin transporter), member 2 |
| chr10_+_11207438 | 0.10 |
ENST00000609692.1
ENST00000354897.3 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr4_+_41614909 | 0.10 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr6_-_87804815 | 0.10 |
ENST00000369582.2
|
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr16_-_745946 | 0.10 |
ENST00000562563.1
|
FBXL16
|
F-box and leucine-rich repeat protein 16 |
| chr17_+_79679299 | 0.10 |
ENST00000331531.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr12_-_56753858 | 0.10 |
ENST00000314128.4
ENST00000557235.1 ENST00000418572.2 |
STAT2
|
signal transducer and activator of transcription 2, 113kDa |
| chr4_-_65275162 | 0.10 |
ENST00000381210.3
ENST00000507440.1 |
TECRL
|
trans-2,3-enoyl-CoA reductase-like |
| chr3_-_151034734 | 0.10 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chr5_-_137878887 | 0.10 |
ENST00000507939.1
ENST00000572514.1 ENST00000499810.2 ENST00000360541.5 |
ETF1
|
eukaryotic translation termination factor 1 |
| chr17_+_79679369 | 0.10 |
ENST00000350690.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr12_+_93963590 | 0.10 |
ENST00000340600.2
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr8_-_70745575 | 0.10 |
ENST00000524945.1
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1 |
| chr14_+_42076765 | 0.09 |
ENST00000298119.4
|
LRFN5
|
leucine rich repeat and fibronectin type III domain containing 5 |
| chr22_+_18121562 | 0.09 |
ENST00000355028.3
|
BCL2L13
|
BCL2-like 13 (apoptosis facilitator) |
| chr16_-_58328923 | 0.09 |
ENST00000567164.1
ENST00000219301.4 ENST00000569727.1 |
PRSS54
|
protease, serine, 54 |
| chr19_+_49547099 | 0.09 |
ENST00000301408.6
|
CGB5
|
chorionic gonadotropin, beta polypeptide 5 |
| chr11_+_67798114 | 0.09 |
ENST00000453471.2
ENST00000528492.1 ENST00000526339.1 ENST00000525419.1 |
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr19_-_8008533 | 0.09 |
ENST00000597926.1
|
TIMM44
|
translocase of inner mitochondrial membrane 44 homolog (yeast) |
| chr4_+_24797085 | 0.09 |
ENST00000382120.3
|
SOD3
|
superoxide dismutase 3, extracellular |
| chr15_+_78441663 | 0.09 |
ENST00000299518.2
ENST00000558554.1 ENST00000557826.1 ENST00000561279.1 ENST00000559186.1 ENST00000560770.1 ENST00000559881.1 ENST00000559205.1 ENST00000441490.2 |
IDH3A
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr17_+_79670386 | 0.09 |
ENST00000333676.3
ENST00000571730.1 ENST00000541223.1 |
MRPL12
SLC25A10
SLC25A10
|
mitochondrial ribosomal protein L12 Mitochondrial dicarboxylate carrier; Uncharacterized protein; cDNA FLJ60124, highly similar to Mitochondrial dicarboxylate carrier solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr17_-_2614927 | 0.09 |
ENST00000435359.1
|
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr2_+_210288760 | 0.08 |
ENST00000199940.6
|
MAP2
|
microtubule-associated protein 2 |
| chr16_+_3704822 | 0.08 |
ENST00000414110.2
|
DNASE1
|
deoxyribonuclease I |
| chr11_+_12766583 | 0.08 |
ENST00000361985.2
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr7_+_129007964 | 0.08 |
ENST00000460109.1
ENST00000474594.1 ENST00000446212.1 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr11_-_64013663 | 0.08 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr19_-_10446449 | 0.08 |
ENST00000592439.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chrX_-_150067272 | 0.07 |
ENST00000355149.3
ENST00000437787.2 |
CD99L2
|
CD99 molecule-like 2 |
| chr10_-_101841588 | 0.07 |
ENST00000370418.3
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr22_-_27620603 | 0.07 |
ENST00000418271.1
ENST00000444114.1 |
RP5-1172A22.1
|
RP5-1172A22.1 |
| chr1_+_45140360 | 0.07 |
ENST00000418644.1
ENST00000458657.2 ENST00000441519.1 ENST00000535358.1 ENST00000445071.1 |
C1orf228
|
chromosome 1 open reading frame 228 |
| chr16_-_58328870 | 0.07 |
ENST00000543437.1
|
PRSS54
|
protease, serine, 54 |
| chr17_-_56358287 | 0.07 |
ENST00000225275.3
ENST00000340482.3 |
MPO
|
myeloperoxidase |
| chr6_-_76072719 | 0.07 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr10_-_51623203 | 0.07 |
ENST00000444743.1
ENST00000374065.3 ENST00000374064.3 ENST00000260867.4 |
TIMM23
|
translocase of inner mitochondrial membrane 23 homolog (yeast) |
| chr16_+_30937213 | 0.07 |
ENST00000427128.1
|
FBXL19
|
F-box and leucine-rich repeat protein 19 |
| chr14_+_21498360 | 0.07 |
ENST00000321760.6
ENST00000460647.2 ENST00000530140.2 ENST00000472458.1 |
TPPP2
|
tubulin polymerization-promoting protein family member 2 |
| chr9_+_113431059 | 0.07 |
ENST00000416899.2
|
MUSK
|
muscle, skeletal, receptor tyrosine kinase |
| chr11_-_64014379 | 0.06 |
ENST00000309318.3
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr3_+_138340067 | 0.06 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr22_-_30783075 | 0.06 |
ENST00000215798.6
|
RNF215
|
ring finger protein 215 |
| chr5_+_149569520 | 0.06 |
ENST00000230671.2
ENST00000524041.1 |
SLC6A7
|
solute carrier family 6 (neurotransmitter transporter), member 7 |
| chrX_-_150067173 | 0.06 |
ENST00000370377.3
ENST00000320893.6 |
CD99L2
|
CD99 molecule-like 2 |
| chr4_-_151936865 | 0.06 |
ENST00000535741.1
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr12_-_108955070 | 0.06 |
ENST00000228284.3
ENST00000546611.1 |
SART3
|
squamous cell carcinoma antigen recognized by T cells 3 |
| chr3_+_128968437 | 0.06 |
ENST00000314797.6
|
COPG1
|
coatomer protein complex, subunit gamma 1 |
| chr2_-_26467557 | 0.06 |
ENST00000380649.3
|
HADHA
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit |
| chr20_+_47538357 | 0.05 |
ENST00000371917.4
|
ARFGEF2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) |
| chr20_+_57875457 | 0.05 |
ENST00000337938.2
ENST00000311585.7 ENST00000371028.2 |
EDN3
|
endothelin 3 |
| chr15_+_43985725 | 0.05 |
ENST00000413453.2
|
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr4_-_122872909 | 0.05 |
ENST00000379645.3
|
TRPC3
|
transient receptor potential cation channel, subfamily C, member 3 |
| chr16_-_28937027 | 0.05 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr16_+_30675654 | 0.05 |
ENST00000287468.5
ENST00000395073.2 |
FBRS
|
fibrosin |
| chr9_-_115983641 | 0.05 |
ENST00000238256.3
|
FKBP15
|
FK506 binding protein 15, 133kDa |
| chr12_-_63544718 | 0.05 |
ENST00000299178.2
|
AVPR1A
|
arginine vasopressin receptor 1A |
| chr1_-_154164534 | 0.05 |
ENST00000271850.7
ENST00000368530.2 |
TPM3
|
tropomyosin 3 |
| chr9_+_113431029 | 0.05 |
ENST00000189978.5
ENST00000374448.4 ENST00000374440.3 |
MUSK
|
muscle, skeletal, receptor tyrosine kinase |
| chrY_-_15591485 | 0.05 |
ENST00000382896.4
ENST00000537580.1 ENST00000540140.1 ENST00000545955.1 ENST00000538878.1 |
UTY
|
ubiquitously transcribed tetratricopeptide repeat containing, Y-linked |
| chr1_-_20987889 | 0.05 |
ENST00000415136.2
|
DDOST
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr7_-_81399438 | 0.05 |
ENST00000222390.5
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr11_-_790060 | 0.05 |
ENST00000330106.4
|
CEND1
|
cell cycle exit and neuronal differentiation 1 |
| chr2_-_121223697 | 0.05 |
ENST00000593290.1
|
FLJ14816
|
long intergenic non-protein coding RNA 1101 |
| chr3_+_138340049 | 0.05 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr14_+_22320634 | 0.05 |
ENST00000390435.1
|
TRAV8-3
|
T cell receptor alpha variable 8-3 |
| chr12_-_22487618 | 0.05 |
ENST00000404299.3
ENST00000396037.4 |
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr19_+_8509842 | 0.04 |
ENST00000325495.4
ENST00000600092.1 ENST00000594907.1 ENST00000596984.1 ENST00000601645.1 |
HNRNPM
|
heterogeneous nuclear ribonucleoprotein M |
| chr3_+_9745510 | 0.04 |
ENST00000383831.3
|
CPNE9
|
copine family member IX |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR4A2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.5 | 1.5 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.3 | 1.0 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.3 | 1.0 | GO:1900075 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.3 | 1.8 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.3 | 0.9 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.3 | 0.9 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.2 | 1.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.2 | 0.5 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.1 | 0.6 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 0.2 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 | 0.3 | GO:1902356 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.1 | 0.3 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 0.6 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 0.2 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.4 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 0.8 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 0.2 | GO:0070676 | regulation of multivesicular body size(GO:0010796) intralumenal vesicle formation(GO:0070676) |
| 0.1 | 0.2 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.1 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.0 | GO:0021699 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.2 | GO:2000211 | negative regulation of cellular amino acid metabolic process(GO:0045763) regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.6 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.3 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.5 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.4 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.2 | GO:1903286 | regulation of potassium ion import(GO:1903286) |
| 0.0 | 0.5 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.7 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.7 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.2 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.4 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.8 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.7 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.5 | GO:0035855 | positive regulation of lamellipodium assembly(GO:0010592) megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.4 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.3 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.3 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.0 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.0 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.1 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.2 | 0.9 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 0.5 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 1.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.3 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 1.0 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 0.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.3 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.5 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 1.3 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 0.4 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.4 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.4 | GO:0031095 | spectrin-associated cytoskeleton(GO:0014731) platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.2 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.2 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.1 | GO:0032279 | axonemal microtubule(GO:0005879) asymmetric synapse(GO:0032279) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.3 | 1.8 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.4 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.1 | 0.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.7 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 1.0 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 1.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.3 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.1 | 0.4 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.2 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.1 | 0.5 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 1.2 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.1 | 0.2 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.1 | 0.6 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 1.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 1.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.3 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.3 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.5 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.3 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.9 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.7 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.0 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 1.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.9 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.1 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.6 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |