Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
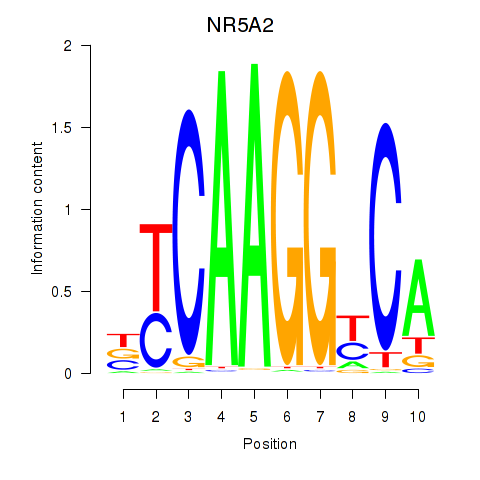
Results for NR5A2
Z-value: 0.67
Transcription factors associated with NR5A2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR5A2
|
ENSG00000116833.9 | nuclear receptor subfamily 5 group A member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR5A2 | hg19_v2_chr1_+_199996702_199996732 | 0.32 | 1.2e-01 | Click! |
Activity profile of NR5A2 motif
Sorted Z-values of NR5A2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_142232071 | 1.00 |
ENST00000390364.3
|
TRBV10-1
|
T cell receptor beta variable 10-1(gene/pseudogene) |
| chr22_+_31518938 | 0.86 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr3_-_156840776 | 0.76 |
ENST00000471357.1
|
LINC00880
|
long intergenic non-protein coding RNA 880 |
| chr15_-_78526855 | 0.69 |
ENST00000541759.1
ENST00000558130.1 |
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr5_+_52285144 | 0.64 |
ENST00000296585.5
|
ITGA2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) |
| chr7_+_89841000 | 0.55 |
ENST00000287908.3
|
STEAP2
|
STEAP family member 2, metalloreductase |
| chr9_+_126131131 | 0.54 |
ENST00000373629.2
|
CRB2
|
crumbs homolog 2 (Drosophila) |
| chr7_+_89841024 | 0.51 |
ENST00000394626.1
|
STEAP2
|
STEAP family member 2, metalloreductase |
| chr14_-_106054659 | 0.48 |
ENST00000390539.2
|
IGHA2
|
immunoglobulin heavy constant alpha 2 (A2m marker) |
| chr8_-_93107696 | 0.47 |
ENST00000436581.2
ENST00000520583.1 ENST00000519061.1 |
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr7_+_142919130 | 0.46 |
ENST00000408947.3
|
TAS2R40
|
taste receptor, type 2, member 40 |
| chr14_-_91526462 | 0.46 |
ENST00000536315.2
|
RPS6KA5
|
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
| chr11_-_790060 | 0.43 |
ENST00000330106.4
|
CEND1
|
cell cycle exit and neuronal differentiation 1 |
| chr2_-_106054952 | 0.42 |
ENST00000336660.5
ENST00000393352.3 ENST00000607522.1 |
FHL2
|
four and a half LIM domains 2 |
| chr4_-_74964904 | 0.40 |
ENST00000508487.2
|
CXCL2
|
chemokine (C-X-C motif) ligand 2 |
| chr6_-_27860956 | 0.39 |
ENST00000359611.2
|
HIST1H2AM
|
histone cluster 1, H2am |
| chr3_-_183735731 | 0.37 |
ENST00000334444.6
|
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr1_+_84609944 | 0.37 |
ENST00000370685.3
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr19_-_40931891 | 0.35 |
ENST00000357949.4
|
SERTAD1
|
SERTA domain containing 1 |
| chr4_-_74904398 | 0.35 |
ENST00000296026.4
|
CXCL3
|
chemokine (C-X-C motif) ligand 3 |
| chrX_-_107225600 | 0.34 |
ENST00000302917.1
|
TEX13B
|
testis expressed 13B |
| chr6_-_27806117 | 0.34 |
ENST00000330180.2
|
HIST1H2AK
|
histone cluster 1, H2ak |
| chr8_-_93107827 | 0.34 |
ENST00000520724.1
ENST00000518844.1 |
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr7_+_2443202 | 0.33 |
ENST00000258711.6
|
CHST12
|
carbohydrate (chondroitin 4) sulfotransferase 12 |
| chr19_-_18995029 | 0.32 |
ENST00000596048.1
|
CERS1
|
ceramide synthase 1 |
| chr10_-_49482907 | 0.32 |
ENST00000374201.3
ENST00000407470.4 |
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr1_+_16083154 | 0.32 |
ENST00000375771.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr9_-_100459639 | 0.30 |
ENST00000375128.4
|
XPA
|
xeroderma pigmentosum, complementation group A |
| chr20_+_42544782 | 0.30 |
ENST00000423191.2
ENST00000372999.1 |
TOX2
|
TOX high mobility group box family member 2 |
| chr1_+_26146397 | 0.28 |
ENST00000374303.2
ENST00000533762.1 ENST00000529116.1 ENST00000474295.1 ENST00000488327.2 ENST00000472643.1 ENST00000526894.1 ENST00000524618.1 ENST00000374307.5 |
MTFR1L
|
mitochondrial fission regulator 1-like |
| chr11_+_7618413 | 0.27 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chrX_-_70474910 | 0.27 |
ENST00000373988.1
ENST00000373998.1 |
ZMYM3
|
zinc finger, MYM-type 3 |
| chr19_-_17356697 | 0.27 |
ENST00000291442.3
|
NR2F6
|
nuclear receptor subfamily 2, group F, member 6 |
| chr16_+_31366536 | 0.27 |
ENST00000562522.1
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chrX_-_70329118 | 0.26 |
ENST00000374188.3
|
IL2RG
|
interleukin 2 receptor, gamma |
| chr11_-_68518910 | 0.26 |
ENST00000544963.1
ENST00000443940.2 |
MTL5
|
metallothionein-like 5, testis-specific (tesmin) |
| chr3_-_122512619 | 0.26 |
ENST00000383659.1
ENST00000306103.2 |
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1 |
| chr1_+_26147319 | 0.26 |
ENST00000374300.3
|
MTFR1L
|
mitochondrial fission regulator 1-like |
| chr14_-_106174960 | 0.26 |
ENST00000390547.2
|
IGHA1
|
immunoglobulin heavy constant alpha 1 |
| chr6_-_26235206 | 0.26 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr1_+_167063282 | 0.26 |
ENST00000361200.2
|
DUSP27
|
dual specificity phosphatase 27 (putative) |
| chr6_+_27782788 | 0.26 |
ENST00000359465.4
|
HIST1H2BM
|
histone cluster 1, H2bm |
| chr15_-_74043816 | 0.25 |
ENST00000379822.4
|
C15orf59
|
chromosome 15 open reading frame 59 |
| chr16_+_56623433 | 0.25 |
ENST00000570176.1
|
MT3
|
metallothionein 3 |
| chr22_+_42372764 | 0.25 |
ENST00000396426.3
ENST00000406029.1 |
SEPT3
|
septin 3 |
| chr12_-_10151773 | 0.25 |
ENST00000298527.6
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1, member B |
| chr1_-_151798546 | 0.25 |
ENST00000356728.6
|
RORC
|
RAR-related orphan receptor C |
| chr2_-_165424973 | 0.25 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr11_+_7597639 | 0.25 |
ENST00000533792.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr9_-_14314518 | 0.24 |
ENST00000397581.2
|
NFIB
|
nuclear factor I/B |
| chr19_+_1248547 | 0.24 |
ENST00000586757.1
ENST00000300952.2 |
MIDN
|
midnolin |
| chr17_-_7080801 | 0.24 |
ENST00000572879.1
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr11_+_59480899 | 0.24 |
ENST00000300150.7
|
STX3
|
syntaxin 3 |
| chr3_-_46506563 | 0.24 |
ENST00000231751.4
|
LTF
|
lactotransferrin |
| chr7_+_35756186 | 0.24 |
ENST00000430518.1
|
AC018647.3
|
AC018647.3 |
| chr11_+_77899920 | 0.23 |
ENST00000528910.1
ENST00000529308.1 |
USP35
|
ubiquitin specific peptidase 35 |
| chr3_-_46506358 | 0.23 |
ENST00000417439.1
ENST00000431944.1 |
LTF
|
lactotransferrin |
| chr6_-_29648887 | 0.23 |
ENST00000376883.1
|
ZFP57
|
ZFP57 zinc finger protein |
| chr7_+_35756092 | 0.23 |
ENST00000458087.3
|
AC018647.3
|
AC018647.3 |
| chr10_-_75634326 | 0.23 |
ENST00000322635.3
ENST00000444854.2 ENST00000423381.1 ENST00000322680.3 ENST00000394762.2 |
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr3_-_58652523 | 0.23 |
ENST00000489857.1
ENST00000358781.2 |
FAM3D
|
family with sequence similarity 3, member D |
| chr10_-_75634260 | 0.23 |
ENST00000372765.1
ENST00000351293.3 |
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr13_-_99174252 | 0.23 |
ENST00000376547.3
|
STK24
|
serine/threonine kinase 24 |
| chr4_+_41614720 | 0.23 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr9_-_14313641 | 0.23 |
ENST00000380953.1
|
NFIB
|
nuclear factor I/B |
| chr3_+_51705222 | 0.23 |
ENST00000457573.1
ENST00000341333.5 ENST00000412249.1 ENST00000425781.1 ENST00000415259.1 ENST00000395057.1 ENST00000416589.1 |
TEX264
|
testis expressed 264 |
| chr2_+_177134134 | 0.23 |
ENST00000249442.6
ENST00000392529.2 ENST00000443241.1 |
MTX2
|
metaxin 2 |
| chr9_-_14314066 | 0.22 |
ENST00000397575.3
|
NFIB
|
nuclear factor I/B |
| chr1_+_231114795 | 0.22 |
ENST00000310256.2
ENST00000366658.2 ENST00000450711.1 ENST00000435927.1 |
ARV1
|
ARV1 homolog (S. cerevisiae) |
| chr2_-_3584430 | 0.22 |
ENST00000438482.1
ENST00000422961.1 |
AC108488.4
|
AC108488.4 |
| chr7_+_29519662 | 0.22 |
ENST00000424025.2
ENST00000439711.2 ENST00000421775.2 |
CHN2
|
chimerin 2 |
| chr12_+_51632666 | 0.21 |
ENST00000604900.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr2_+_177134201 | 0.21 |
ENST00000452865.1
|
MTX2
|
metaxin 2 |
| chr11_-_82746587 | 0.21 |
ENST00000528379.1
ENST00000534103.1 |
RAB30
|
RAB30, member RAS oncogene family |
| chr3_+_38035610 | 0.21 |
ENST00000465644.1
|
VILL
|
villin-like |
| chr16_+_8814563 | 0.21 |
ENST00000425191.2
ENST00000569156.1 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr3_+_179322481 | 0.21 |
ENST00000259037.3
|
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr1_+_26146674 | 0.21 |
ENST00000525713.1
ENST00000374301.3 |
MTFR1L
|
mitochondrial fission regulator 1-like |
| chr9_+_75229616 | 0.20 |
ENST00000340019.3
|
TMC1
|
transmembrane channel-like 1 |
| chr11_-_33913708 | 0.20 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr19_-_1513188 | 0.20 |
ENST00000330475.4
|
ADAMTSL5
|
ADAMTS-like 5 |
| chr1_+_152881014 | 0.20 |
ENST00000368764.3
ENST00000392667.2 |
IVL
|
involucrin |
| chr7_-_38389573 | 0.20 |
ENST00000390344.2
|
TRGV5
|
T cell receptor gamma variable 5 |
| chr12_-_111358372 | 0.20 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr10_-_75634219 | 0.20 |
ENST00000305762.7
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr2_-_111230393 | 0.20 |
ENST00000447537.2
ENST00000413601.2 |
LIMS3L
LIMS3L
|
LIM and senescent cell antigen-like-containing domain protein 3; Uncharacterized protein; cDNA FLJ59124, highly similar to Particularly interesting newCys-His protein; cDNA, FLJ79109, highly similar to Particularly interesting newCys-His protein LIM and senescent cell antigen-like domains 3-like |
| chr14_-_74226961 | 0.20 |
ENST00000286523.5
ENST00000435371.1 |
ELMSAN1
|
ELM2 and Myb/SANT-like domain containing 1 |
| chr18_-_19284724 | 0.20 |
ENST00000580981.1
ENST00000289119.2 |
ABHD3
|
abhydrolase domain containing 3 |
| chr16_+_3194211 | 0.20 |
ENST00000428155.1
|
CASP16
|
caspase 16, apoptosis-related cysteine peptidase (putative) |
| chr5_+_141016969 | 0.19 |
ENST00000518856.1
|
RELL2
|
RELT-like 2 |
| chr3_-_49377499 | 0.19 |
ENST00000265560.4
|
USP4
|
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr9_+_131873227 | 0.19 |
ENST00000358994.4
ENST00000455292.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr8_+_67405755 | 0.19 |
ENST00000521495.1
|
C8orf46
|
chromosome 8 open reading frame 46 |
| chr14_+_37667118 | 0.19 |
ENST00000556615.1
ENST00000327441.7 ENST00000536774.1 |
MIPOL1
|
mirror-image polydactyly 1 |
| chr22_+_29702996 | 0.19 |
ENST00000406549.3
ENST00000360113.2 ENST00000341313.6 ENST00000403764.1 ENST00000471961.1 ENST00000407854.1 |
GAS2L1
|
growth arrest-specific 2 like 1 |
| chr1_-_231114542 | 0.19 |
ENST00000522821.1
ENST00000366661.4 ENST00000366662.4 ENST00000414259.1 ENST00000522399.1 |
TTC13
|
tetratricopeptide repeat domain 13 |
| chr14_-_75330537 | 0.19 |
ENST00000556084.2
ENST00000556489.2 ENST00000445876.1 |
PROX2
|
prospero homeobox 2 |
| chr2_-_88427568 | 0.19 |
ENST00000393750.3
ENST00000295834.3 |
FABP1
|
fatty acid binding protein 1, liver |
| chr6_-_144329531 | 0.19 |
ENST00000429150.1
ENST00000392309.1 ENST00000416623.1 ENST00000392307.1 |
PLAGL1
|
pleiomorphic adenoma gene-like 1 |
| chr14_+_37667193 | 0.19 |
ENST00000539062.2
|
MIPOL1
|
mirror-image polydactyly 1 |
| chr12_+_122667658 | 0.19 |
ENST00000339777.4
ENST00000425921.1 |
LRRC43
|
leucine rich repeat containing 43 |
| chr10_+_23728198 | 0.18 |
ENST00000376495.3
|
OTUD1
|
OTU domain containing 1 |
| chr7_+_129906660 | 0.18 |
ENST00000222481.4
|
CPA2
|
carboxypeptidase A2 (pancreatic) |
| chr1_+_150337144 | 0.18 |
ENST00000539519.1
ENST00000369067.3 ENST00000369068.4 |
RPRD2
|
regulation of nuclear pre-mRNA domain containing 2 |
| chr16_-_57809015 | 0.18 |
ENST00000540079.2
ENST00000569222.1 |
KIFC3
|
kinesin family member C3 |
| chr11_-_63993690 | 0.18 |
ENST00000394546.2
ENST00000541278.1 |
TRPT1
|
tRNA phosphotransferase 1 |
| chr1_-_202129704 | 0.18 |
ENST00000476061.1
ENST00000544762.1 ENST00000467283.1 ENST00000464870.1 ENST00000435759.2 ENST00000486116.1 ENST00000543735.1 ENST00000308986.5 ENST00000477625.1 |
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chrX_-_63005405 | 0.18 |
ENST00000374878.1
ENST00000437457.2 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr19_+_15052301 | 0.18 |
ENST00000248072.3
|
OR7C2
|
olfactory receptor, family 7, subfamily C, member 2 |
| chr1_-_22109682 | 0.18 |
ENST00000400301.1
ENST00000532737.1 |
USP48
|
ubiquitin specific peptidase 48 |
| chr5_-_41213607 | 0.18 |
ENST00000337836.5
ENST00000433294.1 |
C6
|
complement component 6 |
| chr11_-_63993601 | 0.18 |
ENST00000545812.1
ENST00000394547.3 ENST00000317459.6 |
TRPT1
|
tRNA phosphotransferase 1 |
| chr11_-_86666427 | 0.18 |
ENST00000531380.1
|
FZD4
|
frizzled family receptor 4 |
| chr1_+_33231268 | 0.18 |
ENST00000373480.1
|
KIAA1522
|
KIAA1522 |
| chr20_+_30598231 | 0.18 |
ENST00000300415.8
ENST00000262659.8 |
CCM2L
|
cerebral cavernous malformation 2-like |
| chr4_+_74735102 | 0.18 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr1_+_32084641 | 0.18 |
ENST00000373706.5
|
HCRTR1
|
hypocretin (orexin) receptor 1 |
| chr12_-_117537240 | 0.18 |
ENST00000392545.4
ENST00000541210.1 ENST00000335209.7 |
TESC
|
tescalcin |
| chr20_-_36156125 | 0.17 |
ENST00000397135.1
ENST00000397137.1 |
BLCAP
|
bladder cancer associated protein |
| chr19_+_41284121 | 0.17 |
ENST00000594800.1
ENST00000357052.2 ENST00000602173.1 |
RAB4B
|
RAB4B, member RAS oncogene family |
| chr7_+_150782945 | 0.17 |
ENST00000463381.1
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr1_+_162039558 | 0.17 |
ENST00000530878.1
ENST00000361897.5 |
NOS1AP
|
nitric oxide synthase 1 (neuronal) adaptor protein |
| chrX_-_153714994 | 0.17 |
ENST00000369660.4
|
UBL4A
|
ubiquitin-like 4A |
| chr10_-_76995769 | 0.17 |
ENST00000372538.3
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chrX_-_46618490 | 0.17 |
ENST00000328306.4
|
SLC9A7
|
solute carrier family 9, subfamily A (NHE7, cation proton antiporter 7), member 7 |
| chr19_-_291365 | 0.17 |
ENST00000591572.1
ENST00000269812.3 ENST00000434325.2 |
PPAP2C
|
phosphatidic acid phosphatase type 2C |
| chr10_-_104179682 | 0.17 |
ENST00000406432.1
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr20_+_68306 | 0.17 |
ENST00000382410.2
|
DEFB125
|
defensin, beta 125 |
| chr14_+_91580777 | 0.17 |
ENST00000525393.2
ENST00000428926.2 ENST00000517362.1 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr10_-_76995675 | 0.17 |
ENST00000469299.1
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr19_-_55574538 | 0.17 |
ENST00000415061.3
|
RDH13
|
retinol dehydrogenase 13 (all-trans/9-cis) |
| chr7_-_30029367 | 0.17 |
ENST00000242059.5
|
SCRN1
|
secernin 1 |
| chr17_+_30469473 | 0.17 |
ENST00000333942.6
ENST00000358365.3 ENST00000583994.1 ENST00000545287.2 |
RHOT1
|
ras homolog family member T1 |
| chr15_+_73735490 | 0.17 |
ENST00000331090.6
ENST00000560581.1 |
C15orf60
|
chromosome 15 open reading frame 60 |
| chr3_+_9745510 | 0.17 |
ENST00000383831.3
|
CPNE9
|
copine family member IX |
| chr1_-_161993616 | 0.16 |
ENST00000294794.3
|
OLFML2B
|
olfactomedin-like 2B |
| chr10_-_75401500 | 0.16 |
ENST00000359322.4
|
MYOZ1
|
myozenin 1 |
| chr22_+_38035459 | 0.16 |
ENST00000357436.4
|
SH3BP1
|
SH3-domain binding protein 1 |
| chr20_-_44259893 | 0.16 |
ENST00000326000.1
|
WFDC9
|
WAP four-disulfide core domain 9 |
| chr22_-_37608325 | 0.16 |
ENST00000328544.3
|
SSTR3
|
somatostatin receptor 3 |
| chr22_+_19710468 | 0.16 |
ENST00000366425.3
|
GP1BB
|
glycoprotein Ib (platelet), beta polypeptide |
| chr4_+_154387480 | 0.16 |
ENST00000409663.3
ENST00000440693.1 ENST00000409959.3 |
KIAA0922
|
KIAA0922 |
| chr19_+_14017116 | 0.16 |
ENST00000589606.1
|
CC2D1A
|
coiled-coil and C2 domain containing 1A |
| chr16_-_84273304 | 0.16 |
ENST00000308251.4
ENST00000568181.1 |
KCNG4
|
potassium voltage-gated channel, subfamily G, member 4 |
| chr19_-_51529849 | 0.16 |
ENST00000600362.1
ENST00000453757.3 ENST00000601671.1 |
KLK11
|
kallikrein-related peptidase 11 |
| chr3_-_125775629 | 0.16 |
ENST00000383598.2
|
SLC41A3
|
solute carrier family 41, member 3 |
| chr14_+_91580708 | 0.16 |
ENST00000518868.1
|
C14orf159
|
chromosome 14 open reading frame 159 |
| chr3_+_185046676 | 0.16 |
ENST00000428617.1
ENST00000443863.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr2_+_204571375 | 0.16 |
ENST00000374478.4
|
CD28
|
CD28 molecule |
| chr7_-_30029574 | 0.16 |
ENST00000426154.1
ENST00000421434.1 ENST00000434476.2 |
SCRN1
|
secernin 1 |
| chr15_+_83776137 | 0.16 |
ENST00000322019.9
|
TM6SF1
|
transmembrane 6 superfamily member 1 |
| chr2_+_120189422 | 0.16 |
ENST00000306406.4
|
TMEM37
|
transmembrane protein 37 |
| chr5_+_149569520 | 0.16 |
ENST00000230671.2
ENST00000524041.1 |
SLC6A7
|
solute carrier family 6 (neurotransmitter transporter), member 7 |
| chr22_+_38201114 | 0.16 |
ENST00000340857.2
|
H1F0
|
H1 histone family, member 0 |
| chr8_+_37553261 | 0.15 |
ENST00000331569.4
|
ZNF703
|
zinc finger protein 703 |
| chr4_+_175204818 | 0.15 |
ENST00000503780.1
|
CEP44
|
centrosomal protein 44kDa |
| chr7_+_75027418 | 0.15 |
ENST00000447409.2
|
TRIM73
|
tripartite motif containing 73 |
| chr6_+_107349392 | 0.15 |
ENST00000443043.1
ENST00000405204.2 ENST00000311381.5 |
C6orf203
|
chromosome 6 open reading frame 203 |
| chr8_+_94752349 | 0.15 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr3_-_38691119 | 0.15 |
ENST00000333535.4
ENST00000413689.1 ENST00000443581.1 ENST00000425664.1 ENST00000451551.2 |
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit |
| chr5_-_134914673 | 0.15 |
ENST00000512158.1
|
CXCL14
|
chemokine (C-X-C motif) ligand 14 |
| chr1_+_27719148 | 0.15 |
ENST00000374024.3
|
GPR3
|
G protein-coupled receptor 3 |
| chr3_+_16926441 | 0.15 |
ENST00000418129.2
ENST00000396755.2 |
PLCL2
|
phospholipase C-like 2 |
| chr13_-_46679144 | 0.15 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr9_-_14313893 | 0.15 |
ENST00000380921.3
ENST00000380959.3 |
NFIB
|
nuclear factor I/B |
| chr2_-_218770168 | 0.15 |
ENST00000413554.1
|
TNS1
|
tensin 1 |
| chr13_-_46679185 | 0.15 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr12_-_100656134 | 0.15 |
ENST00000548313.1
|
DEPDC4
|
DEP domain containing 4 |
| chr20_+_37434329 | 0.15 |
ENST00000299824.1
ENST00000373331.2 |
PPP1R16B
|
protein phosphatase 1, regulatory subunit 16B |
| chr6_-_105850937 | 0.15 |
ENST00000369110.3
|
PREP
|
prolyl endopeptidase |
| chr14_+_91581011 | 0.15 |
ENST00000523894.1
ENST00000522322.1 ENST00000523771.1 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr11_+_7506713 | 0.15 |
ENST00000329293.3
ENST00000534244.1 |
OLFML1
|
olfactomedin-like 1 |
| chr9_-_94124171 | 0.15 |
ENST00000422391.2
ENST00000375731.4 ENST00000303617.5 |
AUH
|
AU RNA binding protein/enoyl-CoA hydratase |
| chr12_+_49740700 | 0.14 |
ENST00000549441.2
ENST00000395069.3 |
DNAJC22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr9_+_133978190 | 0.14 |
ENST00000372312.3
|
AIF1L
|
allograft inflammatory factor 1-like |
| chr16_-_58328870 | 0.14 |
ENST00000543437.1
|
PRSS54
|
protease, serine, 54 |
| chr3_-_49158312 | 0.14 |
ENST00000398892.3
ENST00000453664.1 ENST00000398888.2 |
USP19
|
ubiquitin specific peptidase 19 |
| chr6_-_52860171 | 0.14 |
ENST00000370963.4
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr16_+_3704822 | 0.14 |
ENST00000414110.2
|
DNASE1
|
deoxyribonuclease I |
| chr22_+_29702572 | 0.14 |
ENST00000407647.2
ENST00000416823.1 ENST00000428622.1 |
GAS2L1
|
growth arrest-specific 2 like 1 |
| chrX_-_49056635 | 0.14 |
ENST00000472598.1
ENST00000538567.1 ENST00000479808.1 ENST00000263233.4 |
SYP
|
synaptophysin |
| chr15_+_67835517 | 0.14 |
ENST00000395476.2
|
MAP2K5
|
mitogen-activated protein kinase kinase 5 |
| chr12_+_48513009 | 0.14 |
ENST00000359794.5
ENST00000551339.1 ENST00000395233.2 ENST00000548345.1 |
PFKM
|
phosphofructokinase, muscle |
| chr11_-_68519026 | 0.14 |
ENST00000255087.5
|
MTL5
|
metallothionein-like 5, testis-specific (tesmin) |
| chr12_+_48513570 | 0.14 |
ENST00000551804.1
|
PFKM
|
phosphofructokinase, muscle |
| chr16_+_11038345 | 0.14 |
ENST00000409790.1
|
CLEC16A
|
C-type lectin domain family 16, member A |
| chr17_-_9683238 | 0.14 |
ENST00000571771.1
|
DHRS7C
|
dehydrogenase/reductase (SDR family) member 7C |
| chr2_+_113403434 | 0.14 |
ENST00000272542.3
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1 |
| chr7_+_29237354 | 0.14 |
ENST00000546235.1
|
CHN2
|
chimerin 2 |
| chr19_-_36643329 | 0.14 |
ENST00000589154.1
|
COX7A1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr16_-_66952742 | 0.14 |
ENST00000565235.2
ENST00000568632.1 ENST00000565796.1 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr17_-_39093672 | 0.14 |
ENST00000209718.3
ENST00000436344.3 ENST00000485751.1 |
KRT23
|
keratin 23 (histone deacetylase inducible) |
| chr1_+_180897269 | 0.13 |
ENST00000367587.1
|
KIAA1614
|
KIAA1614 |
| chr14_+_71648152 | 0.13 |
ENST00000561794.1
|
RP6-91H8.2
|
RP6-91H8.2 |
| chr3_+_186358148 | 0.13 |
ENST00000382134.3
ENST00000265029.3 |
FETUB
|
fetuin B |
| chr1_-_149889382 | 0.13 |
ENST00000369145.1
ENST00000369146.3 |
SV2A
|
synaptic vesicle glycoprotein 2A |
| chr4_-_21699380 | 0.13 |
ENST00000382148.3
|
KCNIP4
|
Kv channel interacting protein 4 |
| chr3_+_186358200 | 0.13 |
ENST00000382136.3
|
FETUB
|
fetuin B |
| chr1_-_201342364 | 0.13 |
ENST00000236918.7
ENST00000367317.4 ENST00000367315.2 ENST00000360372.4 |
TNNT2
|
troponin T type 2 (cardiac) |
| chr5_-_135231516 | 0.13 |
ENST00000274520.1
|
IL9
|
interleukin 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR5A2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.2 | 0.5 | GO:2000308 | iron assimilation(GO:0033212) iron assimilation by chelation and transport(GO:0033214) positive regulation of bone mineralization involved in bone maturation(GO:1900159) negative regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000308) |
| 0.1 | 0.4 | GO:0036146 | cellular response to mycotoxin(GO:0036146) |
| 0.1 | 1.1 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.1 | 0.8 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.3 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.1 | 0.5 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 | 0.5 | GO:0014028 | notochord formation(GO:0014028) |
| 0.1 | 0.4 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.1 | 0.3 | GO:2000296 | cadmium ion homeostasis(GO:0055073) negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.1 | 0.4 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.2 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.1 | 0.2 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 0.1 | 0.3 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.1 | 0.7 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.3 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.1 | 0.2 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.1 | 0.2 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.2 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.2 | GO:1903947 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.1 | 0.2 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.1 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.1 | 0.4 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.2 | GO:1904448 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.1 | 0.3 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.3 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.1 | GO:0003213 | cardiac right atrium morphogenesis(GO:0003213) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0050894 | determination of affect(GO:0050894) |
| 0.0 | 0.2 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.0 | 0.2 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.2 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.2 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.2 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.2 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.3 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.9 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.2 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.2 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.2 | GO:1902513 | regulation of organelle transport along microtubule(GO:1902513) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.1 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.0 | 0.1 | GO:0003064 | regulation of heart rate by hormone(GO:0003064) |
| 0.0 | 0.2 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.1 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.0 | 0.2 | GO:0038169 | hormone-mediated apoptotic signaling pathway(GO:0008628) somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.1 | GO:0097187 | dentinogenesis(GO:0097187) |
| 0.0 | 0.2 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.7 | GO:0014733 | regulation of skeletal muscle adaptation(GO:0014733) |
| 0.0 | 0.3 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0036378 | alkaloid catabolic process(GO:0009822) calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.0 | 0.1 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.2 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.0 | 0.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.7 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.2 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.0 | 0.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.1 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.1 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.1 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.0 | 0.1 | GO:0019542 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.0 | 0.2 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 0.3 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.4 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) positive regulation of endodeoxyribonuclease activity(GO:0032079) regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.2 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.5 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.2 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.0 | 0.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.2 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.3 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.1 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.7 | GO:0006536 | glutamate metabolic process(GO:0006536) |
| 0.0 | 0.5 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.0 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.0 | GO:0015728 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.0 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0002863 | positive regulation of inflammatory response to antigenic stimulus(GO:0002863) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.0 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.0 | 0.3 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 0.2 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.0 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.1 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 0.1 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.0 | 0.1 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.3 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.5 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.0 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.0 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.0 | 0.2 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0071746 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 0.1 | 0.6 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.1 | 0.3 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.1 | 0.5 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.1 | 0.2 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 0.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.2 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 0.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.3 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.2 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 1.1 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.0 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.9 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.7 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.1 | 0.7 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.1 | 0.6 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.3 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 0.9 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.1 | 0.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 0.4 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.1 | 0.3 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 0.3 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.1 | 0.3 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.1 | 0.2 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.1 | 0.2 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.1 | 0.2 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.1 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.0 | 0.7 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.0 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.2 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 0.8 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.2 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.3 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.5 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.2 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.1 | GO:0047783 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.0 | 0.2 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.2 | GO:0050543 | icosatetraenoic acid binding(GO:0050543) arachidonic acid binding(GO:0050544) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.2 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.0 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.6 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.2 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.1 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.9 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 1.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |