Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
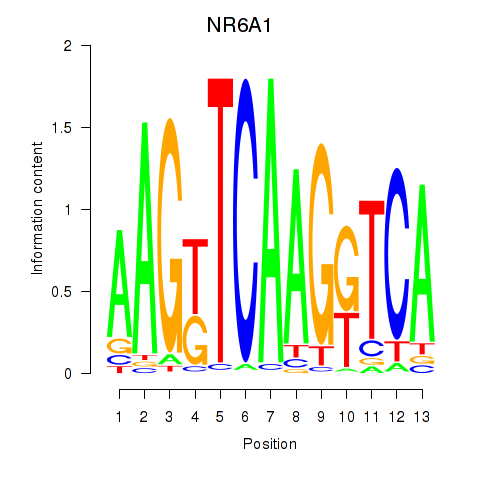
Results for NR6A1
Z-value: 0.64
Transcription factors associated with NR6A1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR6A1
|
ENSG00000148200.12 | nuclear receptor subfamily 6 group A member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR6A1 | hg19_v2_chr9_-_127533582_127533615 | 0.61 | 1.2e-03 | Click! |
Activity profile of NR6A1 motif
Sorted Z-values of NR6A1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_76071805 | 0.96 |
ENST00000539311.1
|
FLVCR2
|
feline leukemia virus subgroup C cellular receptor family, member 2 |
| chr19_-_43586820 | 0.75 |
ENST00000406487.1
|
PSG2
|
pregnancy specific beta-1-glycoprotein 2 |
| chr19_+_41699135 | 0.71 |
ENST00000542619.1
ENST00000600561.1 |
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr19_+_41698927 | 0.62 |
ENST00000310054.4
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr21_+_43639211 | 0.58 |
ENST00000450121.1
ENST00000398449.3 ENST00000361802.2 |
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr17_-_46799872 | 0.51 |
ENST00000290294.3
|
PRAC1
|
prostate cancer susceptibility candidate 1 |
| chr6_-_160166218 | 0.51 |
ENST00000537657.1
|
SOD2
|
superoxide dismutase 2, mitochondrial |
| chr11_-_35287243 | 0.49 |
ENST00000464522.2
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr7_-_142232071 | 0.47 |
ENST00000390364.3
|
TRBV10-1
|
T cell receptor beta variable 10-1(gene/pseudogene) |
| chr12_+_60058458 | 0.43 |
ENST00000548610.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr10_+_116853201 | 0.41 |
ENST00000527407.1
|
ATRNL1
|
attractin-like 1 |
| chr1_+_78511586 | 0.41 |
ENST00000370759.3
|
GIPC2
|
GIPC PDZ domain containing family, member 2 |
| chr11_-_108464465 | 0.41 |
ENST00000525344.1
|
EXPH5
|
exophilin 5 |
| chr9_-_38069208 | 0.40 |
ENST00000377707.3
ENST00000377700.4 |
SHB
|
Src homology 2 domain containing adaptor protein B |
| chr7_-_108168580 | 0.40 |
ENST00000453085.1
|
PNPLA8
|
patatin-like phospholipase domain containing 8 |
| chr2_+_201994208 | 0.39 |
ENST00000440180.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr15_+_76629064 | 0.38 |
ENST00000290759.4
|
ISL2
|
ISL LIM homeobox 2 |
| chr1_-_161102421 | 0.38 |
ENST00000490843.2
ENST00000368006.3 ENST00000392188.1 ENST00000545495.1 |
DEDD
|
death effector domain containing |
| chr11_+_34643600 | 0.37 |
ENST00000530286.1
ENST00000533754.1 |
EHF
|
ets homologous factor |
| chr10_+_81107271 | 0.37 |
ENST00000448165.1
|
PPIF
|
peptidylprolyl isomerase F |
| chr19_-_43382142 | 0.37 |
ENST00000597058.1
|
PSG1
|
pregnancy specific beta-1-glycoprotein 1 |
| chr1_+_212738676 | 0.36 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr1_-_161102367 | 0.36 |
ENST00000464113.1
|
DEDD
|
death effector domain containing |
| chr6_+_84743436 | 0.35 |
ENST00000257776.4
|
MRAP2
|
melanocortin 2 receptor accessory protein 2 |
| chr16_-_75498308 | 0.35 |
ENST00000569540.1
|
TMEM170A
|
transmembrane protein 170A |
| chr11_-_11747257 | 0.35 |
ENST00000601641.1
|
AC131935.1
|
AC131935.1 |
| chr17_-_56494713 | 0.34 |
ENST00000407977.2
|
RNF43
|
ring finger protein 43 |
| chr10_+_124030819 | 0.33 |
ENST00000260723.4
ENST00000368994.2 |
BTBD16
|
BTB (POZ) domain containing 16 |
| chr11_+_66188475 | 0.33 |
ENST00000311034.2
|
NPAS4
|
neuronal PAS domain protein 4 |
| chr3_-_139396560 | 0.33 |
ENST00000514703.1
ENST00000511444.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr13_-_114898016 | 0.33 |
ENST00000542651.1
ENST00000334062.7 |
RASA3
|
RAS p21 protein activator 3 |
| chr1_-_202679535 | 0.32 |
ENST00000367268.4
|
SYT2
|
synaptotagmin II |
| chr5_+_140749803 | 0.31 |
ENST00000576222.1
|
PCDHGB3
|
protocadherin gamma subfamily B, 3 |
| chr8_-_102275131 | 0.31 |
ENST00000523121.1
|
KB-1410C5.2
|
KB-1410C5.2 |
| chr7_+_150521691 | 0.31 |
ENST00000493429.1
|
AOC1
|
amine oxidase, copper containing 1 |
| chr8_-_141645645 | 0.30 |
ENST00000519980.1
ENST00000220592.5 |
AGO2
|
argonaute RISC catalytic component 2 |
| chr19_+_53761545 | 0.30 |
ENST00000341702.3
|
VN1R2
|
vomeronasal 1 receptor 2 |
| chr17_-_37309480 | 0.29 |
ENST00000539608.1
|
PLXDC1
|
plexin domain containing 1 |
| chr8_-_17555164 | 0.28 |
ENST00000297488.6
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr16_-_75498553 | 0.27 |
ENST00000569276.1
ENST00000357613.4 ENST00000561878.1 ENST00000566980.1 ENST00000567194.1 |
TMEM170A
RP11-77K12.1
|
transmembrane protein 170A Uncharacterized protein |
| chr4_-_159094194 | 0.27 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr8_-_27472198 | 0.27 |
ENST00000519472.1
ENST00000523589.1 ENST00000522413.1 ENST00000523396.1 ENST00000560366.1 |
CLU
|
clusterin |
| chr19_+_15160130 | 0.27 |
ENST00000427043.3
|
CASP14
|
caspase 14, apoptosis-related cysteine peptidase |
| chr7_+_37960163 | 0.26 |
ENST00000199448.4
ENST00000559325.1 ENST00000423717.1 |
EPDR1
|
ependymin related 1 |
| chr19_-_43422019 | 0.26 |
ENST00000402603.4
ENST00000594375.1 |
PSG6
|
pregnancy specific beta-1-glycoprotein 6 |
| chr5_-_156486120 | 0.26 |
ENST00000522693.1
|
HAVCR1
|
hepatitis A virus cellular receptor 1 |
| chr17_-_37308824 | 0.26 |
ENST00000415163.1
ENST00000441877.1 ENST00000444911.2 |
PLXDC1
|
plexin domain containing 1 |
| chr3_+_37284824 | 0.26 |
ENST00000431105.1
|
GOLGA4
|
golgin A4 |
| chr2_-_25564750 | 0.25 |
ENST00000321117.5
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha |
| chr1_+_107682629 | 0.25 |
ENST00000370074.4
ENST00000370073.2 ENST00000370071.2 ENST00000542803.1 ENST00000370061.3 ENST00000370072.3 ENST00000370070.2 |
NTNG1
|
netrin G1 |
| chr14_+_22670455 | 0.25 |
ENST00000390460.1
|
TRAV26-2
|
T cell receptor alpha variable 26-2 |
| chr17_-_71410794 | 0.25 |
ENST00000424778.1
|
SDK2
|
sidekick cell adhesion molecule 2 |
| chr11_+_111789580 | 0.25 |
ENST00000278601.5
|
C11orf52
|
chromosome 11 open reading frame 52 |
| chr9_-_14314066 | 0.24 |
ENST00000397575.3
|
NFIB
|
nuclear factor I/B |
| chr17_+_66509019 | 0.24 |
ENST00000585981.1
ENST00000589480.1 ENST00000585815.1 |
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr10_+_116853091 | 0.24 |
ENST00000526946.1
|
ATRNL1
|
attractin-like 1 |
| chr9_+_75136717 | 0.24 |
ENST00000297784.5
|
TMC1
|
transmembrane channel-like 1 |
| chr3_+_114012819 | 0.24 |
ENST00000383671.3
|
TIGIT
|
T cell immunoreceptor with Ig and ITIM domains |
| chr13_+_23755099 | 0.24 |
ENST00000537476.1
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
| chr19_-_58864848 | 0.24 |
ENST00000263100.3
|
A1BG
|
alpha-1-B glycoprotein |
| chr12_-_91574142 | 0.24 |
ENST00000547937.1
|
DCN
|
decorin |
| chr2_+_234296792 | 0.23 |
ENST00000409813.3
|
DGKD
|
diacylglycerol kinase, delta 130kDa |
| chr1_-_161207875 | 0.23 |
ENST00000512372.1
ENST00000437437.2 ENST00000442691.2 ENST00000412844.2 ENST00000428574.2 ENST00000505005.1 ENST00000508740.1 ENST00000508387.1 ENST00000504010.1 ENST00000511676.1 ENST00000502985.1 ENST00000367981.3 ENST00000515621.1 ENST00000511944.1 ENST00000511748.1 ENST00000367984.4 ENST00000367985.3 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr3_-_156840776 | 0.23 |
ENST00000471357.1
|
LINC00880
|
long intergenic non-protein coding RNA 880 |
| chr12_-_24737089 | 0.23 |
ENST00000483544.1
|
LINC00477
|
long intergenic non-protein coding RNA 477 |
| chr17_-_79817091 | 0.23 |
ENST00000570907.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr12_+_69186125 | 0.23 |
ENST00000399333.3
|
AC124890.1
|
HCG1774533, isoform CRA_a; PRO2268; Uncharacterized protein |
| chr13_+_23755054 | 0.23 |
ENST00000218867.3
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
| chr12_-_109219937 | 0.22 |
ENST00000546697.1
|
SSH1
|
slingshot protein phosphatase 1 |
| chr16_+_31366455 | 0.22 |
ENST00000268296.4
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr2_-_175711133 | 0.22 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr11_+_62009653 | 0.22 |
ENST00000244926.3
|
SCGB1D2
|
secretoglobin, family 1D, member 2 |
| chr21_+_10862622 | 0.21 |
ENST00000302092.5
ENST00000559480.1 |
IGHV1OR21-1
|
immunoglobulin heavy variable 1/OR21-1 (non-functional) |
| chr17_+_7942424 | 0.21 |
ENST00000573359.1
|
ALOX15B
|
arachidonate 15-lipoxygenase, type B |
| chr16_-_420338 | 0.21 |
ENST00000450882.1
ENST00000441883.1 ENST00000447696.1 ENST00000389675.2 |
MRPL28
|
mitochondrial ribosomal protein L28 |
| chr17_-_2614927 | 0.20 |
ENST00000435359.1
|
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr17_+_7531281 | 0.20 |
ENST00000575729.1
ENST00000340624.5 |
SHBG
|
sex hormone-binding globulin |
| chr11_+_65190245 | 0.20 |
ENST00000499732.1
ENST00000501122.2 ENST00000601801.1 |
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
| chr7_+_72349920 | 0.20 |
ENST00000395270.1
ENST00000446813.1 ENST00000257622.4 |
POM121
|
POM121 transmembrane nucleoporin |
| chr12_-_7261772 | 0.20 |
ENST00000545280.1
ENST00000543933.1 ENST00000545337.1 ENST00000544702.1 ENST00000266542.4 |
C1RL
|
complement component 1, r subcomponent-like |
| chr20_+_31619454 | 0.20 |
ENST00000349552.1
|
BPIFB6
|
BPI fold containing family B, member 6 |
| chr11_-_104480019 | 0.20 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr6_-_31697977 | 0.19 |
ENST00000375787.2
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr16_-_2836606 | 0.19 |
ENST00000571674.1
ENST00000293851.5 ENST00000576886.1 |
PRSS33
|
protease, serine, 33 |
| chr16_-_420514 | 0.19 |
ENST00000199706.8
|
MRPL28
|
mitochondrial ribosomal protein L28 |
| chr19_+_8953269 | 0.19 |
ENST00000305625.2
|
MBD3L1
|
methyl-CpG binding domain protein 3-like 1 |
| chr8_-_72268889 | 0.19 |
ENST00000388742.4
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr1_-_184943610 | 0.19 |
ENST00000367511.3
|
FAM129A
|
family with sequence similarity 129, member A |
| chr22_+_40390930 | 0.19 |
ENST00000333407.6
|
FAM83F
|
family with sequence similarity 83, member F |
| chr22_+_45680822 | 0.18 |
ENST00000216211.4
ENST00000396082.2 |
UPK3A
|
uroplakin 3A |
| chr19_-_7293942 | 0.18 |
ENST00000341500.5
ENST00000302850.5 |
INSR
|
insulin receptor |
| chr5_+_55147205 | 0.18 |
ENST00000396836.2
ENST00000396834.1 ENST00000447346.2 ENST00000359040.5 |
IL31RA
|
interleukin 31 receptor A |
| chr15_-_20170354 | 0.18 |
ENST00000338912.5
|
IGHV1OR15-9
|
immunoglobulin heavy variable 1/OR15-9 (non-functional) |
| chrX_+_54466829 | 0.18 |
ENST00000375151.4
|
TSR2
|
TSR2, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr1_-_112032284 | 0.18 |
ENST00000414219.1
|
ADORA3
|
adenosine A3 receptor |
| chr11_+_113185533 | 0.18 |
ENST00000393020.1
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chr3_-_139396801 | 0.18 |
ENST00000413939.2
ENST00000339837.5 ENST00000512391.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr12_+_9980069 | 0.18 |
ENST00000354855.3
ENST00000324214.4 ENST00000279544.3 |
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr11_-_124190184 | 0.17 |
ENST00000357438.2
|
OR8D2
|
olfactory receptor, family 8, subfamily D, member 2 |
| chr18_+_11751493 | 0.17 |
ENST00000269162.5
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr3_-_139396853 | 0.17 |
ENST00000406164.1
ENST00000406824.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr1_+_45212051 | 0.17 |
ENST00000372222.3
|
KIF2C
|
kinesin family member 2C |
| chr9_-_111619239 | 0.17 |
ENST00000374667.3
|
ACTL7B
|
actin-like 7B |
| chr18_+_616672 | 0.17 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr18_+_616711 | 0.17 |
ENST00000579494.1
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr17_-_8198636 | 0.16 |
ENST00000577745.1
ENST00000579192.1 ENST00000396278.1 |
SLC25A35
|
solute carrier family 25, member 35 |
| chr16_-_30122717 | 0.16 |
ENST00000566613.1
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr5_-_138725594 | 0.16 |
ENST00000302125.8
|
MZB1
|
marginal zone B and B1 cell-specific protein |
| chr1_-_161207953 | 0.16 |
ENST00000367982.4
|
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr5_-_138725560 | 0.16 |
ENST00000412103.2
ENST00000457570.2 |
MZB1
|
marginal zone B and B1 cell-specific protein |
| chr5_-_41213607 | 0.16 |
ENST00000337836.5
ENST00000433294.1 |
C6
|
complement component 6 |
| chr19_+_51376505 | 0.16 |
ENST00000325321.3
ENST00000600690.1 ENST00000391810.2 ENST00000358049.4 |
KLK2
|
kallikrein-related peptidase 2 |
| chr18_+_11751466 | 0.16 |
ENST00000535121.1
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chrX_-_118827113 | 0.16 |
ENST00000394617.2
|
SEPT6
|
septin 6 |
| chr9_-_140115775 | 0.15 |
ENST00000391553.1
ENST00000392827.1 |
RNF208
|
ring finger protein 208 |
| chr22_-_24181174 | 0.15 |
ENST00000318109.7
ENST00000406855.3 ENST00000404056.1 ENST00000476077.1 |
DERL3
|
derlin 3 |
| chr1_-_161208013 | 0.15 |
ENST00000515452.1
ENST00000367983.4 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr9_-_39239171 | 0.15 |
ENST00000358144.2
|
CNTNAP3
|
contactin associated protein-like 3 |
| chr18_+_23713808 | 0.15 |
ENST00000415576.2
ENST00000343848.6 ENST00000308268.6 |
PSMA8
|
proteasome (prosome, macropain) subunit, alpha type, 8 |
| chr17_-_27503770 | 0.15 |
ENST00000533112.1
|
MYO18A
|
myosin XVIIIA |
| chr2_-_26467465 | 0.15 |
ENST00000457468.2
|
HADHA
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit |
| chr10_+_114710516 | 0.15 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr5_-_135231516 | 0.15 |
ENST00000274520.1
|
IL9
|
interleukin 9 |
| chr1_+_45212074 | 0.15 |
ENST00000372217.1
|
KIF2C
|
kinesin family member 2C |
| chr12_+_10366016 | 0.15 |
ENST00000546017.1
ENST00000535576.1 ENST00000539170.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr9_-_14313641 | 0.15 |
ENST00000380953.1
|
NFIB
|
nuclear factor I/B |
| chrX_+_103031758 | 0.14 |
ENST00000303958.2
ENST00000361621.2 |
PLP1
|
proteolipid protein 1 |
| chr7_+_94536898 | 0.14 |
ENST00000433360.1
ENST00000340694.4 ENST00000424654.1 |
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr7_-_56160625 | 0.14 |
ENST00000446428.1
ENST00000432123.1 ENST00000452681.2 ENST00000537360.1 |
PHKG1
|
phosphorylase kinase, gamma 1 (muscle) |
| chr3_-_113464906 | 0.14 |
ENST00000477813.1
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr13_+_111766897 | 0.14 |
ENST00000317133.5
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr8_-_72268968 | 0.14 |
ENST00000388740.3
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr11_-_108464321 | 0.14 |
ENST00000265843.4
|
EXPH5
|
exophilin 5 |
| chr7_+_102004322 | 0.14 |
ENST00000496391.1
|
PRKRIP1
|
PRKR interacting protein 1 (IL11 inducible) |
| chr3_+_113465866 | 0.13 |
ENST00000273398.3
ENST00000538620.1 ENST00000496747.1 ENST00000475322.1 |
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
| chr17_+_32612687 | 0.13 |
ENST00000305869.3
|
CCL11
|
chemokine (C-C motif) ligand 11 |
| chr11_-_73720122 | 0.13 |
ENST00000426995.2
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr5_+_169780485 | 0.13 |
ENST00000377360.4
|
KCNIP1
|
Kv channel interacting protein 1 |
| chr22_-_50217981 | 0.13 |
ENST00000457780.2
|
BRD1
|
bromodomain containing 1 |
| chr1_+_29213584 | 0.13 |
ENST00000343067.4
ENST00000356093.2 ENST00000398863.2 ENST00000373800.3 ENST00000349460.4 |
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr3_+_73045936 | 0.13 |
ENST00000356692.5
ENST00000488810.1 ENST00000394284.3 ENST00000295862.9 ENST00000495566.1 |
PPP4R2
|
protein phosphatase 4, regulatory subunit 2 |
| chr21_+_42694732 | 0.13 |
ENST00000398646.3
|
FAM3B
|
family with sequence similarity 3, member B |
| chr22_+_42372764 | 0.13 |
ENST00000396426.3
ENST00000406029.1 |
SEPT3
|
septin 3 |
| chr6_-_25874440 | 0.13 |
ENST00000361703.6
ENST00000397060.4 |
SLC17A3
|
solute carrier family 17 (organic anion transporter), member 3 |
| chr19_-_43269809 | 0.13 |
ENST00000406636.3
ENST00000404209.4 ENST00000306511.4 |
PSG8
|
pregnancy specific beta-1-glycoprotein 8 |
| chr9_-_14314518 | 0.13 |
ENST00000397581.2
|
NFIB
|
nuclear factor I/B |
| chrY_+_16636354 | 0.13 |
ENST00000339174.5
|
NLGN4Y
|
neuroligin 4, Y-linked |
| chr2_-_60780607 | 0.13 |
ENST00000537768.1
ENST00000335712.6 ENST00000356842.4 |
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr9_+_976964 | 0.12 |
ENST00000190165.2
|
DMRT3
|
doublesex and mab-3 related transcription factor 3 |
| chr2_+_26568965 | 0.12 |
ENST00000260585.7
ENST00000447170.1 |
EPT1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) |
| chr19_-_8373173 | 0.12 |
ENST00000537716.2
ENST00000301458.5 |
CD320
|
CD320 molecule |
| chr17_-_40729681 | 0.12 |
ENST00000590760.1
ENST00000587209.1 ENST00000393795.3 ENST00000253789.5 |
PSMC3IP
|
PSMC3 interacting protein |
| chr7_-_69062391 | 0.12 |
ENST00000436600.2
|
RP5-942I16.1
|
RP5-942I16.1 |
| chr6_+_31620191 | 0.12 |
ENST00000375918.2
ENST00000375920.4 |
APOM
|
apolipoprotein M |
| chr8_-_27468842 | 0.12 |
ENST00000523500.1
|
CLU
|
clusterin |
| chr17_-_39093672 | 0.12 |
ENST00000209718.3
ENST00000436344.3 ENST00000485751.1 |
KRT23
|
keratin 23 (histone deacetylase inducible) |
| chr17_-_7531121 | 0.12 |
ENST00000573566.1
ENST00000269298.5 |
SAT2
|
spermidine/spermine N1-acetyltransferase family member 2 |
| chr19_-_22193706 | 0.12 |
ENST00000597040.1
|
ZNF208
|
zinc finger protein 208 |
| chrX_+_103031421 | 0.12 |
ENST00000433491.1
ENST00000418604.1 ENST00000443502.1 |
PLP1
|
proteolipid protein 1 |
| chr12_+_50017184 | 0.12 |
ENST00000548825.2
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) |
| chr2_+_54558004 | 0.12 |
ENST00000405749.1
ENST00000398634.2 ENST00000447328.1 |
C2orf73
|
chromosome 2 open reading frame 73 |
| chr5_-_146258291 | 0.12 |
ENST00000394411.4
ENST00000453001.1 |
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr6_+_25652432 | 0.12 |
ENST00000377961.2
|
SCGN
|
secretagogin, EF-hand calcium binding protein |
| chr2_+_152266604 | 0.11 |
ENST00000430328.2
|
RIF1
|
RAP1 interacting factor homolog (yeast) |
| chr8_-_145701718 | 0.11 |
ENST00000377317.4
|
FOXH1
|
forkhead box H1 |
| chr9_-_21305312 | 0.11 |
ENST00000259555.4
|
IFNA5
|
interferon, alpha 5 |
| chr19_-_43244610 | 0.11 |
ENST00000595140.1
ENST00000327495.5 |
PSG3
|
pregnancy specific beta-1-glycoprotein 3 |
| chr22_-_50218452 | 0.11 |
ENST00000216267.8
|
BRD1
|
bromodomain containing 1 |
| chr14_-_106471723 | 0.11 |
ENST00000390595.2
|
IGHV1-3
|
immunoglobulin heavy variable 1-3 |
| chr16_-_84538218 | 0.11 |
ENST00000562447.1
ENST00000565765.1 ENST00000535580.1 ENST00000343629.6 |
TLDC1
|
TBC/LysM-associated domain containing 1 |
| chr5_+_72143988 | 0.11 |
ENST00000506351.2
|
TNPO1
|
transportin 1 |
| chr11_-_78052923 | 0.11 |
ENST00000340149.2
|
GAB2
|
GRB2-associated binding protein 2 |
| chr2_+_152266392 | 0.10 |
ENST00000444746.2
ENST00000453091.2 ENST00000428287.2 ENST00000433166.2 ENST00000420714.3 ENST00000243326.5 ENST00000414861.2 |
RIF1
|
RAP1 interacting factor homolog (yeast) |
| chr8_-_16043780 | 0.10 |
ENST00000445506.2
|
MSR1
|
macrophage scavenger receptor 1 |
| chr3_-_139396787 | 0.10 |
ENST00000296202.7
ENST00000509291.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr17_-_15587602 | 0.10 |
ENST00000416464.2
ENST00000578237.1 ENST00000581200.1 |
TRIM16
|
tripartite motif containing 16 |
| chr19_+_11455900 | 0.10 |
ENST00000588790.1
|
CCDC159
|
coiled-coil domain containing 159 |
| chr4_-_170948361 | 0.10 |
ENST00000393702.3
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr10_-_76995675 | 0.10 |
ENST00000469299.1
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr14_-_106539557 | 0.10 |
ENST00000390599.2
|
IGHV1-8
|
immunoglobulin heavy variable 1-8 |
| chr1_-_161207986 | 0.10 |
ENST00000506209.1
ENST00000367980.2 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr1_+_92417716 | 0.10 |
ENST00000402388.1
|
BRDT
|
bromodomain, testis-specific |
| chr7_+_31569365 | 0.10 |
ENST00000454513.1
ENST00000451887.2 |
CCDC129
|
coiled-coil domain containing 129 |
| chr11_-_33743952 | 0.10 |
ENST00000534312.1
|
CD59
|
CD59 molecule, complement regulatory protein |
| chr7_-_75401513 | 0.10 |
ENST00000005180.4
|
CCL26
|
chemokine (C-C motif) ligand 26 |
| chr7_-_95225768 | 0.10 |
ENST00000005178.5
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr10_+_11207438 | 0.10 |
ENST00000609692.1
ENST00000354897.3 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chrX_-_46187069 | 0.10 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr6_+_28109703 | 0.10 |
ENST00000457389.2
ENST00000330236.6 |
ZKSCAN8
|
zinc finger with KRAB and SCAN domains 8 |
| chr16_-_21170762 | 0.10 |
ENST00000261383.3
ENST00000415178.1 |
DNAH3
|
dynein, axonemal, heavy chain 3 |
| chrX_+_56100757 | 0.10 |
ENST00000433279.1
|
AL353698.1
|
Uncharacterized protein |
| chr9_-_122131696 | 0.10 |
ENST00000373964.2
ENST00000265922.3 |
BRINP1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 1 |
| chr10_-_50396425 | 0.10 |
ENST00000374148.1
|
C10orf128
|
chromosome 10 open reading frame 128 |
| chrX_+_23018058 | 0.10 |
ENST00000327968.5
|
DDX53
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 53 |
| chr11_+_118398178 | 0.10 |
ENST00000302783.4
ENST00000539546.1 |
TTC36
|
tetratricopeptide repeat domain 36 |
| chr19_-_43690674 | 0.10 |
ENST00000342951.6
ENST00000366175.3 |
PSG5
|
pregnancy specific beta-1-glycoprotein 5 |
| chr5_-_176836577 | 0.09 |
ENST00000253496.3
|
F12
|
coagulation factor XII (Hageman factor) |
| chr15_-_22082822 | 0.09 |
ENST00000439682.1
|
POTEB
|
POTE ankyrin domain family, member B |
| chr8_-_87081926 | 0.09 |
ENST00000276616.2
|
PSKH2
|
protein serine kinase H2 |
| chr22_+_23229960 | 0.09 |
ENST00000526893.1
ENST00000532223.2 ENST00000531372.1 |
IGLL5
|
immunoglobulin lambda-like polypeptide 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR6A1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0055099 | detection of endogenous stimulus(GO:0009726) response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 0.4 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 0.8 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.4 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.4 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.1 | 0.4 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 0.5 | GO:0001315 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 0.4 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.3 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.1 | 0.5 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.2 | GO:1904719 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.1 | 1.0 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.4 | GO:0061518 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 0.3 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.1 | 0.5 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 0.2 | GO:1904799 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.1 | 0.4 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.3 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.1 | 1.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.1 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.0 | 0.2 | GO:1990535 | transformation of host cell by virus(GO:0019087) neuron projection maintenance(GO:1990535) |
| 0.0 | 0.1 | GO:0035962 | response to interleukin-13(GO:0035962) |
| 0.0 | 0.3 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.3 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.2 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.4 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.0 | 0.3 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:0051039 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 0.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.1 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.0 | 0.1 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.0 | 0.2 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.0 | 0.6 | GO:0051292 | nuclear pore complex assembly(GO:0051292) endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.1 | GO:0050894 | determination of affect(GO:0050894) |
| 0.0 | 0.2 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:0043105 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.0 | 0.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.2 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) myotome development(GO:0061055) |
| 0.0 | 0.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.0 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.0 | 0.0 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.3 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.3 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.0 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.2 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.0 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.0 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 0.0 | GO:1904815 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.7 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0032173 | septin ring(GO:0005940) septin collar(GO:0032173) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.3 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.4 | GO:0097342 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0033011 | perinuclear theca(GO:0033011) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.3 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.0 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0001741 | XY body(GO:0001741) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.2 | 0.6 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 0.8 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.1 | 0.6 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.1 | 0.4 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.3 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 0.3 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 0.3 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 1.0 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.2 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.1 | 0.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.5 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 0.2 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.4 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.3 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.3 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.0 | 0.2 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.1 | GO:0017057 | glucose-6-phosphate dehydrogenase activity(GO:0004345) 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.1 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.3 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.0 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.2 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |