Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for OLIG3_NEUROD2_NEUROG2
Z-value: 0.65
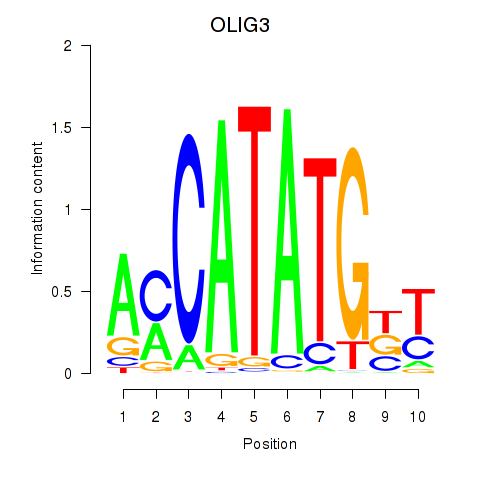
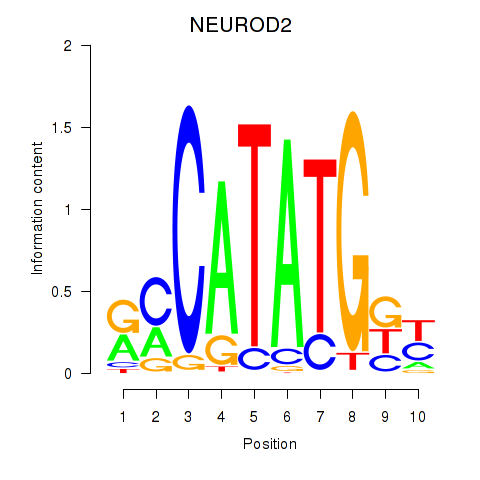
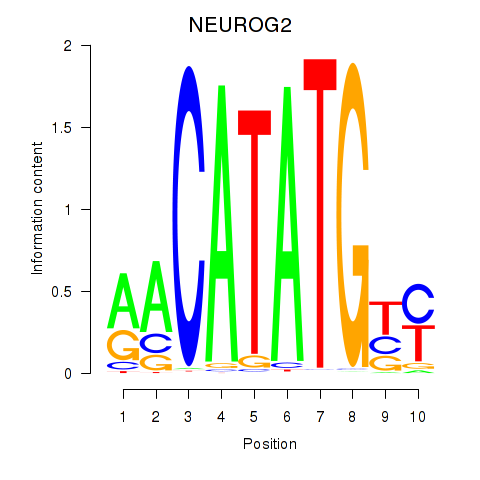
Transcription factors associated with OLIG3_NEUROD2_NEUROG2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OLIG3
|
ENSG00000177468.5 | oligodendrocyte transcription factor 3 |
|
NEUROD2
|
ENSG00000171532.4 | neuronal differentiation 2 |
|
NEUROG2
|
ENSG00000178403.3 | neurogenin 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NEUROG2 | hg19_v2_chr4_-_113437328_113437337 | -0.21 | 3.1e-01 | Click! |
| OLIG3 | hg19_v2_chr6_-_137815524_137815537 | -0.19 | 3.5e-01 | Click! |
| NEUROD2 | hg19_v2_chr17_-_37764128_37764258 | -0.13 | 5.4e-01 | Click! |
Activity profile of OLIG3_NEUROD2_NEUROG2 motif
Sorted Z-values of OLIG3_NEUROD2_NEUROG2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_144354644 | 2.10 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr5_-_111312622 | 1.69 |
ENST00000395634.3
|
NREP
|
neuronal regeneration related protein |
| chr3_-_18480260 | 1.61 |
ENST00000454909.2
|
SATB1
|
SATB homeobox 1 |
| chr1_+_47489240 | 1.58 |
ENST00000371901.3
|
CYP4X1
|
cytochrome P450, family 4, subfamily X, polypeptide 1 |
| chr8_-_93029865 | 1.39 |
ENST00000422361.2
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr3_-_108672742 | 1.18 |
ENST00000261047.3
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr2_+_108994466 | 1.14 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr17_-_58469591 | 1.11 |
ENST00000589335.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr7_-_11871815 | 1.00 |
ENST00000423059.4
|
THSD7A
|
thrombospondin, type I, domain containing 7A |
| chr2_+_108994633 | 0.98 |
ENST00000409309.3
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr3_-_108672609 | 0.98 |
ENST00000393963.3
ENST00000471108.1 |
GUCA1C
|
guanylate cyclase activator 1C |
| chr19_-_11450249 | 0.98 |
ENST00000222120.3
|
RAB3D
|
RAB3D, member RAS oncogene family |
| chr13_+_49551020 | 0.97 |
ENST00000541916.1
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr1_-_186344802 | 0.93 |
ENST00000451586.1
|
TPR
|
translocated promoter region, nuclear basket protein |
| chr14_-_107283278 | 0.87 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr4_+_156587979 | 0.72 |
ENST00000511507.1
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr2_-_188378368 | 0.71 |
ENST00000392365.1
ENST00000435414.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr15_-_98417780 | 0.69 |
ENST00000503874.3
|
LINC00923
|
long intergenic non-protein coding RNA 923 |
| chr17_+_66511540 | 0.68 |
ENST00000588188.2
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr4_+_156587853 | 0.68 |
ENST00000506455.1
ENST00000511108.1 |
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr4_-_159094194 | 0.57 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr10_+_5135981 | 0.56 |
ENST00000380554.3
|
AKR1C3
|
aldo-keto reductase family 1, member C3 |
| chr3_+_49840685 | 0.54 |
ENST00000333323.4
|
FAM212A
|
family with sequence similarity 212, member A |
| chr10_-_7513904 | 0.52 |
ENST00000420395.1
|
RP5-1031D4.2
|
RP5-1031D4.2 |
| chr6_+_3000057 | 0.52 |
ENST00000397717.2
|
NQO2
|
NAD(P)H dehydrogenase, quinone 2 |
| chr9_+_12775011 | 0.48 |
ENST00000319264.3
|
LURAP1L
|
leucine rich adaptor protein 1-like |
| chr12_+_26126681 | 0.47 |
ENST00000542865.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr8_+_21915368 | 0.47 |
ENST00000265800.5
ENST00000517418.1 |
DMTN
|
dematin actin binding protein |
| chr2_-_166930131 | 0.46 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr9_+_72002837 | 0.45 |
ENST00000377216.3
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr5_+_140571902 | 0.44 |
ENST00000239446.4
|
PCDHB10
|
protocadherin beta 10 |
| chr17_+_73629500 | 0.44 |
ENST00000375215.3
|
SMIM5
|
small integral membrane protein 5 |
| chr2_+_177053307 | 0.44 |
ENST00000331462.4
|
HOXD1
|
homeobox D1 |
| chr1_+_180897269 | 0.43 |
ENST00000367587.1
|
KIAA1614
|
KIAA1614 |
| chr7_+_141478242 | 0.41 |
ENST00000247881.2
|
TAS2R4
|
taste receptor, type 2, member 4 |
| chr7_+_116502605 | 0.41 |
ENST00000458284.2
ENST00000490693.1 |
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr11_-_72070206 | 0.41 |
ENST00000544382.1
|
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr1_+_244214577 | 0.41 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr7_-_112758589 | 0.41 |
ENST00000413744.1
ENST00000439551.1 ENST00000441359.1 |
LINC00998
|
long intergenic non-protein coding RNA 998 |
| chr15_+_65843130 | 0.40 |
ENST00000569894.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr11_-_110167352 | 0.39 |
ENST00000533991.1
ENST00000528498.1 ENST00000405097.1 ENST00000528900.1 ENST00000530301.1 ENST00000343115.4 |
RDX
|
radixin |
| chrX_+_118108601 | 0.39 |
ENST00000371628.3
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr19_-_52227221 | 0.39 |
ENST00000222115.1
ENST00000540069.2 |
HAS1
|
hyaluronan synthase 1 |
| chr2_-_183106641 | 0.38 |
ENST00000346717.4
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chrX_+_118108571 | 0.38 |
ENST00000304778.7
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr3_-_197300194 | 0.37 |
ENST00000358186.2
ENST00000431056.1 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr20_+_34802295 | 0.37 |
ENST00000432603.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr2_-_183387430 | 0.37 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr3_-_178984759 | 0.36 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr14_-_69864993 | 0.36 |
ENST00000555373.1
|
ERH
|
enhancer of rudimentary homolog (Drosophila) |
| chr4_-_100575781 | 0.36 |
ENST00000511828.1
|
RP11-766F14.2
|
Protein LOC285556 |
| chr14_+_63671577 | 0.35 |
ENST00000555125.1
|
RHOJ
|
ras homolog family member J |
| chr2_-_165424973 | 0.35 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr9_-_75567962 | 0.35 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chrX_-_125686784 | 0.35 |
ENST00000371126.1
|
DCAF12L1
|
DDB1 and CUL4 associated factor 12-like 1 |
| chr12_+_56862301 | 0.35 |
ENST00000338146.5
|
SPRYD4
|
SPRY domain containing 4 |
| chr6_-_52859046 | 0.34 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr7_-_101212244 | 0.34 |
ENST00000451953.1
ENST00000434537.1 ENST00000437900.1 |
LINC01007
|
long intergenic non-protein coding RNA 1007 |
| chr8_+_7801144 | 0.33 |
ENST00000443676.1
|
ZNF705B
|
zinc finger protein 705B |
| chr17_+_56769924 | 0.33 |
ENST00000461271.1
ENST00000583539.1 ENST00000337432.4 ENST00000421782.2 |
RAD51C
|
RAD51 paralog C |
| chr1_-_21377383 | 0.33 |
ENST00000374935.3
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr6_-_76072719 | 0.32 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr7_-_156685841 | 0.32 |
ENST00000354505.4
ENST00000540390.1 |
LMBR1
|
limb development membrane protein 1 |
| chr6_-_29324054 | 0.32 |
ENST00000543825.1
|
OR5V1
|
olfactory receptor, family 5, subfamily V, member 1 |
| chr11_+_7506713 | 0.32 |
ENST00000329293.3
ENST00000534244.1 |
OLFML1
|
olfactomedin-like 1 |
| chr3_-_195310802 | 0.31 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr3_+_141043050 | 0.31 |
ENST00000509842.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr7_+_114562909 | 0.31 |
ENST00000423503.1
ENST00000427207.1 |
MDFIC
|
MyoD family inhibitor domain containing |
| chr7_+_142031986 | 0.30 |
ENST00000547918.2
|
TRBV7-1
|
T cell receptor beta variable 7-1 (non-functional) |
| chr16_+_81272287 | 0.30 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr2_+_54342574 | 0.30 |
ENST00000303536.4
ENST00000394666.3 |
ACYP2
|
acylphosphatase 2, muscle type |
| chr15_-_71055878 | 0.29 |
ENST00000322954.6
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr17_+_60758814 | 0.29 |
ENST00000579432.1
ENST00000446119.2 |
MRC2
|
mannose receptor, C type 2 |
| chr9_+_5450503 | 0.29 |
ENST00000381573.4
ENST00000381577.3 |
CD274
|
CD274 molecule |
| chr17_+_7211656 | 0.29 |
ENST00000416016.2
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr3_+_119298429 | 0.28 |
ENST00000478927.1
|
ADPRH
|
ADP-ribosylarginine hydrolase |
| chr9_+_2029019 | 0.28 |
ENST00000382194.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_-_183387064 | 0.28 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr6_+_3000195 | 0.28 |
ENST00000338130.2
|
NQO2
|
NAD(P)H dehydrogenase, quinone 2 |
| chr11_-_11374904 | 0.28 |
ENST00000528848.2
|
CSNK2A3
|
casein kinase 2, alpha 3 polypeptide |
| chr2_+_191792376 | 0.28 |
ENST00000409428.1
ENST00000409215.1 |
GLS
|
glutaminase |
| chr11_+_92085707 | 0.28 |
ENST00000525166.1
|
FAT3
|
FAT atypical cadherin 3 |
| chr14_+_22675388 | 0.27 |
ENST00000390461.2
|
TRAV34
|
T cell receptor alpha variable 34 |
| chr11_+_120107344 | 0.27 |
ENST00000260264.4
|
POU2F3
|
POU class 2 homeobox 3 |
| chr16_-_72206034 | 0.27 |
ENST00000537465.1
ENST00000237353.10 |
PMFBP1
|
polyamine modulated factor 1 binding protein 1 |
| chr7_+_134430212 | 0.27 |
ENST00000436461.2
|
CALD1
|
caldesmon 1 |
| chr4_-_116034979 | 0.27 |
ENST00000264363.2
|
NDST4
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 |
| chr12_-_91573132 | 0.26 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr4_-_186733363 | 0.26 |
ENST00000393523.2
ENST00000393528.3 ENST00000449407.2 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr9_-_111775772 | 0.26 |
ENST00000325580.6
ENST00000374593.4 ENST00000374595.4 ENST00000325551.4 |
CTNNAL1
|
catenin (cadherin-associated protein), alpha-like 1 |
| chr1_+_100810575 | 0.25 |
ENST00000542213.1
|
CDC14A
|
cell division cycle 14A |
| chr5_-_171433579 | 0.25 |
ENST00000265094.5
ENST00000393802.2 |
FBXW11
|
F-box and WD repeat domain containing 11 |
| chr6_+_3000218 | 0.25 |
ENST00000380441.1
ENST00000380455.4 ENST00000380454.4 |
NQO2
|
NAD(P)H dehydrogenase, quinone 2 |
| chr12_-_108954933 | 0.25 |
ENST00000431469.2
ENST00000546815.1 |
SART3
|
squamous cell carcinoma antigen recognized by T cells 3 |
| chr10_+_133918175 | 0.25 |
ENST00000298622.4
|
JAKMIP3
|
Janus kinase and microtubule interacting protein 3 |
| chr9_-_125675576 | 0.25 |
ENST00000373659.3
|
ZBTB6
|
zinc finger and BTB domain containing 6 |
| chr15_-_80189380 | 0.25 |
ENST00000258874.3
|
MTHFS
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr20_-_34117447 | 0.25 |
ENST00000246199.2
ENST00000424444.1 ENST00000374345.4 ENST00000444723.1 |
C20orf173
|
chromosome 20 open reading frame 173 |
| chr11_-_134123142 | 0.25 |
ENST00000392595.2
ENST00000341541.3 ENST00000352327.5 ENST00000392594.3 |
THYN1
|
thymocyte nuclear protein 1 |
| chr1_-_21377447 | 0.25 |
ENST00000374937.3
ENST00000264211.8 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr5_-_11588907 | 0.25 |
ENST00000513598.1
ENST00000503622.1 |
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr14_-_38725573 | 0.25 |
ENST00000342213.2
|
CLEC14A
|
C-type lectin domain family 14, member A |
| chr3_-_114477962 | 0.25 |
ENST00000471418.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr11_+_55944094 | 0.25 |
ENST00000312298.1
|
OR5J2
|
olfactory receptor, family 5, subfamily J, member 2 |
| chr15_+_85144217 | 0.24 |
ENST00000540936.1
ENST00000448803.2 ENST00000546275.1 ENST00000546148.1 ENST00000442073.3 ENST00000334141.3 ENST00000358472.3 ENST00000502939.2 ENST00000379358.3 ENST00000327179.6 |
ZSCAN2
|
zinc finger and SCAN domain containing 2 |
| chr2_+_33359687 | 0.24 |
ENST00000402934.1
ENST00000404525.1 ENST00000407925.1 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr19_+_18208603 | 0.24 |
ENST00000262811.6
|
MAST3
|
microtubule associated serine/threonine kinase 3 |
| chr6_-_33714752 | 0.23 |
ENST00000451316.1
|
IP6K3
|
inositol hexakisphosphate kinase 3 |
| chr11_+_134123389 | 0.23 |
ENST00000281182.4
ENST00000537423.1 ENST00000543332.1 ENST00000374752.4 |
ACAD8
|
acyl-CoA dehydrogenase family, member 8 |
| chr9_+_90112117 | 0.23 |
ENST00000358077.5
|
DAPK1
|
death-associated protein kinase 1 |
| chr11_+_118175596 | 0.23 |
ENST00000528600.1
|
CD3E
|
CD3e molecule, epsilon (CD3-TCR complex) |
| chr2_+_33359646 | 0.23 |
ENST00000390003.4
ENST00000418533.2 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr1_+_225600404 | 0.23 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr8_+_86121448 | 0.23 |
ENST00000520225.1
|
E2F5
|
E2F transcription factor 5, p130-binding |
| chr11_+_118175132 | 0.23 |
ENST00000361763.4
|
CD3E
|
CD3e molecule, epsilon (CD3-TCR complex) |
| chr6_-_30712313 | 0.23 |
ENST00000376377.2
ENST00000259874.5 |
IER3
|
immediate early response 3 |
| chr6_-_33714667 | 0.23 |
ENST00000293756.4
|
IP6K3
|
inositol hexakisphosphate kinase 3 |
| chr2_+_136499179 | 0.23 |
ENST00000272638.9
|
UBXN4
|
UBX domain protein 4 |
| chr19_+_35417798 | 0.22 |
ENST00000303586.7
ENST00000439785.1 ENST00000601540.1 |
ZNF30
|
zinc finger protein 30 |
| chr3_-_114477787 | 0.22 |
ENST00000464560.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr12_+_54393880 | 0.22 |
ENST00000303450.4
|
HOXC9
|
homeobox C9 |
| chr8_-_16050142 | 0.22 |
ENST00000536385.1
ENST00000381998.4 |
MSR1
|
macrophage scavenger receptor 1 |
| chr18_+_3448455 | 0.22 |
ENST00000549780.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr10_-_48055018 | 0.22 |
ENST00000426610.2
|
ASAH2C
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2C |
| chr20_+_57226841 | 0.22 |
ENST00000358029.4
ENST00000361830.3 |
STX16
|
syntaxin 16 |
| chrX_+_135278908 | 0.22 |
ENST00000539015.1
ENST00000370683.1 |
FHL1
|
four and a half LIM domains 1 |
| chr16_+_31366455 | 0.22 |
ENST00000268296.4
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr4_-_83719884 | 0.22 |
ENST00000282709.4
ENST00000273908.4 |
SCD5
|
stearoyl-CoA desaturase 5 |
| chr2_+_173600514 | 0.22 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr8_-_16050214 | 0.22 |
ENST00000262101.5
|
MSR1
|
macrophage scavenger receptor 1 |
| chr21_+_17566643 | 0.22 |
ENST00000419952.1
ENST00000445461.2 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr4_-_147043058 | 0.22 |
ENST00000512063.1
ENST00000507726.1 |
RP11-6L6.3
|
long intergenic non-protein coding RNA 1095 |
| chr2_-_113999260 | 0.21 |
ENST00000468980.2
|
PAX8
|
paired box 8 |
| chr1_+_95975672 | 0.21 |
ENST00000440116.2
ENST00000456933.1 |
RP11-286B14.1
|
RP11-286B14.1 |
| chr12_-_102872317 | 0.21 |
ENST00000424202.2
|
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr16_+_12058961 | 0.21 |
ENST00000053243.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr7_-_38403077 | 0.21 |
ENST00000426402.2
|
TRGV2
|
T cell receptor gamma variable 2 |
| chrX_+_47444613 | 0.21 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr14_+_24540046 | 0.20 |
ENST00000397016.2
ENST00000537691.1 ENST00000560356.1 ENST00000558450.1 |
CPNE6
|
copine VI (neuronal) |
| chr12_+_82347498 | 0.20 |
ENST00000550506.1
|
RP11-362A1.1
|
RP11-362A1.1 |
| chr17_-_47925379 | 0.20 |
ENST00000352793.2
ENST00000334568.4 ENST00000398154.1 ENST00000436235.1 ENST00000326219.5 |
TAC4
|
tachykinin 4 (hemokinin) |
| chr16_+_12059050 | 0.20 |
ENST00000396495.3
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr15_-_65503801 | 0.20 |
ENST00000261883.4
|
CILP
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| chr20_+_10199468 | 0.20 |
ENST00000254976.2
ENST00000304886.2 |
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr2_+_173600565 | 0.20 |
ENST00000397081.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr14_+_69865401 | 0.20 |
ENST00000556605.1
ENST00000336643.5 ENST00000031146.4 |
SLC39A9
|
solute carrier family 39, member 9 |
| chr2_+_189839046 | 0.20 |
ENST00000304636.3
ENST00000317840.5 |
COL3A1
|
collagen, type III, alpha 1 |
| chr1_-_55352834 | 0.20 |
ENST00000371269.3
|
DHCR24
|
24-dehydrocholesterol reductase |
| chr15_+_75940218 | 0.20 |
ENST00000308527.5
|
SNX33
|
sorting nexin 33 |
| chr19_+_50194360 | 0.20 |
ENST00000323446.5
ENST00000392518.4 ENST00000598396.1 ENST00000598293.1 ENST00000354199.5 ENST00000405931.2 ENST00000602019.1 |
CPT1C
|
carnitine palmitoyltransferase 1C |
| chr11_-_5537920 | 0.20 |
ENST00000380184.1
|
UBQLNL
|
ubiquilin-like |
| chr19_-_14945933 | 0.20 |
ENST00000322301.3
|
OR7A5
|
olfactory receptor, family 7, subfamily A, member 5 |
| chr13_+_49794474 | 0.20 |
ENST00000218721.1
ENST00000398307.1 |
MLNR
|
motilin receptor |
| chr4_+_71458012 | 0.20 |
ENST00000449493.2
|
AMBN
|
ameloblastin (enamel matrix protein) |
| chr19_-_37019562 | 0.19 |
ENST00000523638.1
|
ZNF260
|
zinc finger protein 260 |
| chr3_-_98241358 | 0.19 |
ENST00000503004.1
ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1
|
claudin domain containing 1 |
| chr14_-_106539557 | 0.19 |
ENST00000390599.2
|
IGHV1-8
|
immunoglobulin heavy variable 1-8 |
| chr8_-_135522425 | 0.19 |
ENST00000521673.1
|
ZFAT
|
zinc finger and AT hook domain containing |
| chr7_+_148287657 | 0.19 |
ENST00000307003.2
|
C7orf33
|
chromosome 7 open reading frame 33 |
| chr5_+_156607829 | 0.19 |
ENST00000422843.3
|
ITK
|
IL2-inducible T-cell kinase |
| chr2_+_71680852 | 0.19 |
ENST00000409762.1
ENST00000409582.3 ENST00000429174.2 ENST00000413539.2 ENST00000258104.3 |
DYSF
|
dysferlin |
| chr4_-_22444733 | 0.19 |
ENST00000508133.1
|
GPR125
|
G protein-coupled receptor 125 |
| chrX_+_78426469 | 0.19 |
ENST00000276077.1
|
GPR174
|
G protein-coupled receptor 174 |
| chr16_-_21877629 | 0.19 |
ENST00000544693.1
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr16_+_2016821 | 0.19 |
ENST00000569210.2
ENST00000569714.1 |
RNF151
|
ring finger protein 151 |
| chr14_+_27342334 | 0.19 |
ENST00000548170.1
ENST00000552926.1 |
RP11-384J4.1
|
RP11-384J4.1 |
| chr16_-_66583701 | 0.19 |
ENST00000527800.1
ENST00000525974.1 ENST00000563369.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr16_+_49407710 | 0.19 |
ENST00000299191.3
|
C16orf78
|
chromosome 16 open reading frame 78 |
| chr8_-_19540086 | 0.18 |
ENST00000332246.6
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr8_-_38239732 | 0.18 |
ENST00000534155.1
ENST00000433384.2 ENST00000317025.8 ENST00000316985.3 |
WHSC1L1
|
Wolf-Hirschhorn syndrome candidate 1-like 1 |
| chr10_-_76995769 | 0.18 |
ENST00000372538.3
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr19_-_1021113 | 0.18 |
ENST00000333175.5
ENST00000356663.3 |
TMEM259
|
transmembrane protein 259 |
| chr17_+_66255310 | 0.18 |
ENST00000448504.2
|
ARSG
|
arylsulfatase G |
| chr10_-_94257512 | 0.18 |
ENST00000371581.5
|
IDE
|
insulin-degrading enzyme |
| chr18_+_77623668 | 0.18 |
ENST00000316249.3
|
KCNG2
|
potassium voltage-gated channel, subfamily G, member 2 |
| chr22_+_44319619 | 0.18 |
ENST00000216180.3
|
PNPLA3
|
patatin-like phospholipase domain containing 3 |
| chr1_-_44818599 | 0.17 |
ENST00000537474.1
|
ERI3
|
ERI1 exoribonuclease family member 3 |
| chr4_+_174818390 | 0.17 |
ENST00000509968.1
ENST00000512263.1 |
RP11-161D15.1
|
RP11-161D15.1 |
| chr5_-_11589131 | 0.17 |
ENST00000511377.1
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr10_-_48416849 | 0.17 |
ENST00000249598.1
|
GDF2
|
growth differentiation factor 2 |
| chr17_+_63133587 | 0.17 |
ENST00000449996.3
ENST00000262406.9 |
RGS9
|
regulator of G-protein signaling 9 |
| chr10_-_76995675 | 0.17 |
ENST00000469299.1
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr5_+_175299743 | 0.17 |
ENST00000502265.1
|
CPLX2
|
complexin 2 |
| chr13_+_28519343 | 0.17 |
ENST00000381026.3
|
ATP5EP2
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit pseudogene 2 |
| chr15_+_74466012 | 0.16 |
ENST00000249842.3
|
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr16_+_53133070 | 0.16 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr4_+_71457970 | 0.16 |
ENST00000322937.6
|
AMBN
|
ameloblastin (enamel matrix protein) |
| chr5_+_135496675 | 0.16 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr16_-_28550348 | 0.16 |
ENST00000324873.6
|
NUPR1
|
nuclear protein, transcriptional regulator, 1 |
| chr5_+_147443534 | 0.16 |
ENST00000398454.1
ENST00000359874.3 ENST00000508733.1 ENST00000256084.7 |
SPINK5
|
serine peptidase inhibitor, Kazal type 5 |
| chr8_-_17752996 | 0.16 |
ENST00000381841.2
ENST00000427924.1 |
FGL1
|
fibrinogen-like 1 |
| chr12_-_108714412 | 0.16 |
ENST00000412676.1
ENST00000550573.1 |
CMKLR1
|
chemokine-like receptor 1 |
| chr11_+_114310237 | 0.16 |
ENST00000539119.1
|
REXO2
|
RNA exonuclease 2 |
| chr9_-_14180778 | 0.16 |
ENST00000380924.1
ENST00000543693.1 |
NFIB
|
nuclear factor I/B |
| chr4_-_150736962 | 0.16 |
ENST00000502345.1
ENST00000510975.1 ENST00000511993.1 |
RP11-526A4.1
|
RP11-526A4.1 |
| chr2_+_179184955 | 0.16 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr16_+_22516172 | 0.16 |
ENST00000543407.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr13_+_111767650 | 0.16 |
ENST00000449979.1
ENST00000370623.3 |
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr11_+_114310164 | 0.16 |
ENST00000544196.1
ENST00000539754.1 ENST00000539275.1 |
REXO2
|
RNA exonuclease 2 |
| chr11_+_121461097 | 0.16 |
ENST00000527934.1
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing |
Network of associatons between targets according to the STRING database.
First level regulatory network of OLIG3_NEUROD2_NEUROG2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0045799 | mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) positive regulation of chromatin assembly or disassembly(GO:0045799) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.2 | 1.4 | GO:0052551 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.2 | 1.0 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.2 | 2.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 0.6 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.1 | 0.4 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.3 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.5 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.3 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.5 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 1.7 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.7 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.3 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.1 | 0.4 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.1 | 0.5 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 2.1 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.3 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.1 | 0.4 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.2 | GO:2000612 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 | 0.2 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.1 | 0.2 | GO:0042560 | folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 | 0.2 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.1 | 1.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.3 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 1.0 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.2 | GO:1902997 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 | 2.1 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.2 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.4 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.5 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.2 | GO:0046104 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.2 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.5 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.1 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.1 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.0 | 1.0 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.2 | GO:1904073 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 | 0.3 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.2 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.0 | 0.4 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.6 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.0 | 0.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.3 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.1 | GO:0001207 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 1.1 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 | 0.2 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.2 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.1 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.2 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.5 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.1 | GO:2001248 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.1 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 0.1 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.1 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.1 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.1 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.1 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.1 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.1 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.1 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.1 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.1 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.0 | 0.2 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.0 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.6 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.9 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.3 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.4 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.3 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.3 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.2 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.0 | 0.0 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.1 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.1 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.0 | 0.1 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.0 | 0.0 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.5 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.0 | GO:0043324 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.1 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.2 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.0 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.0 | 0.1 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.0 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.2 | GO:2000194 | regulation of female gonad development(GO:2000194) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 2.3 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 0.3 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 1.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.9 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.3 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.6 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.9 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.4 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 1.6 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0097182 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.0 | 0.7 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.6 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.3 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.1 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.3 | 1.1 | GO:1904408 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.1 | 1.0 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.6 | GO:0047718 | indanol dehydrogenase activity(GO:0047718) |
| 0.1 | 0.4 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.4 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.1 | 0.4 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.4 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 2.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.2 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.1 | 1.7 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.3 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.1 | 0.3 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 0.2 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.1 | 0.2 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.1 | 0.3 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 0.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.3 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 1.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.1 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.4 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.5 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.7 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.3 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.2 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.2 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 2.0 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.7 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.1 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.0 | 0.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.4 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.5 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.0 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0044390 | ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.0 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.0 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.5 | GO:0030291 | protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.0 | 0.1 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 1.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 2.5 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 2.7 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.2 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.9 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.6 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.7 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.6 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 1.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.7 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.6 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.1 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |