Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
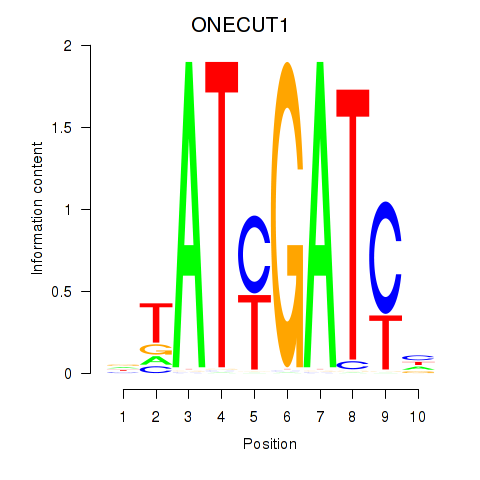
Results for ONECUT1
Z-value: 1.05
Transcription factors associated with ONECUT1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ONECUT1
|
ENSG00000169856.7 | one cut homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ONECUT1 | hg19_v2_chr15_-_53082178_53082209 | -0.33 | 1.0e-01 | Click! |
Activity profile of ONECUT1 motif
Sorted Z-values of ONECUT1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_42792442 | 4.63 |
ENST00000398600.2
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr11_-_72385437 | 4.37 |
ENST00000418754.2
ENST00000542969.2 ENST00000334456.5 |
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated |
| chr21_+_27011899 | 3.56 |
ENST00000425221.2
|
JAM2
|
junctional adhesion molecule 2 |
| chr3_-_79816965 | 3.15 |
ENST00000464233.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr6_+_127898312 | 3.07 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr12_-_76817036 | 2.99 |
ENST00000546946.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr7_-_115670792 | 2.11 |
ENST00000265440.7
ENST00000393485.1 |
TFEC
|
transcription factor EC |
| chr11_+_19799327 | 2.09 |
ENST00000540292.1
|
NAV2
|
neuron navigator 2 |
| chr7_-_115670804 | 2.05 |
ENST00000320239.7
|
TFEC
|
transcription factor EC |
| chr1_-_186649543 | 1.80 |
ENST00000367468.5
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chr20_+_58515417 | 1.67 |
ENST00000360816.3
|
FAM217B
|
family with sequence similarity 217, member B |
| chr11_+_102188272 | 1.58 |
ENST00000532808.1
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr3_-_112693865 | 1.47 |
ENST00000471858.1
ENST00000295863.4 ENST00000308611.3 |
CD200R1
|
CD200 receptor 1 |
| chr4_-_157892055 | 1.44 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr14_-_27066960 | 1.40 |
ENST00000539517.2
|
NOVA1
|
neuro-oncological ventral antigen 1 |
| chr4_-_157892167 | 1.34 |
ENST00000541126.1
|
PDGFC
|
platelet derived growth factor C |
| chr19_+_13135386 | 1.33 |
ENST00000360105.4
ENST00000588228.1 ENST00000591028.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr7_-_69062391 | 1.27 |
ENST00000436600.2
|
RP5-942I16.1
|
RP5-942I16.1 |
| chr19_+_45449301 | 1.25 |
ENST00000591597.1
|
APOC2
|
apolipoprotein C-II |
| chr11_-_118305921 | 1.24 |
ENST00000532619.1
|
RP11-770J1.4
|
RP11-770J1.4 |
| chr7_-_121944491 | 1.14 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr20_-_22559211 | 1.13 |
ENST00000564492.1
|
LINC00261
|
long intergenic non-protein coding RNA 261 |
| chr10_+_60028818 | 1.13 |
ENST00000333926.5
|
CISD1
|
CDGSH iron sulfur domain 1 |
| chr4_-_157892498 | 1.12 |
ENST00000502773.1
|
PDGFC
|
platelet derived growth factor C |
| chr20_-_58515344 | 1.12 |
ENST00000370996.3
|
PPP1R3D
|
protein phosphatase 1, regulatory subunit 3D |
| chr11_+_131240373 | 1.12 |
ENST00000374791.3
ENST00000436745.1 |
NTM
|
neurotrimin |
| chr10_+_114133773 | 1.09 |
ENST00000354655.4
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr5_+_98109322 | 1.07 |
ENST00000513185.1
|
RGMB
|
repulsive guidance molecule family member b |
| chr1_-_151804314 | 1.05 |
ENST00000318247.6
|
RORC
|
RAR-related orphan receptor C |
| chr15_-_64338521 | 1.04 |
ENST00000457488.1
ENST00000558069.1 |
DAPK2
|
death-associated protein kinase 2 |
| chr15_-_89089860 | 1.04 |
ENST00000558413.1
ENST00000564406.1 ENST00000268148.8 |
DET1
|
de-etiolated homolog 1 (Arabidopsis) |
| chr13_-_46679144 | 1.02 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr13_-_46679185 | 1.01 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr12_+_111471828 | 0.99 |
ENST00000261726.6
|
CUX2
|
cut-like homeobox 2 |
| chr3_+_101546827 | 0.97 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr11_-_19223523 | 0.95 |
ENST00000265968.3
|
CSRP3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr11_+_62623544 | 0.94 |
ENST00000377890.2
ENST00000377891.2 ENST00000377889.2 |
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr11_+_62623621 | 0.89 |
ENST00000535296.1
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr1_-_151804222 | 0.86 |
ENST00000392697.3
|
RORC
|
RAR-related orphan receptor C |
| chr8_+_82192501 | 0.85 |
ENST00000297258.6
|
FABP5
|
fatty acid binding protein 5 (psoriasis-associated) |
| chrX_+_122318113 | 0.85 |
ENST00000371264.3
|
GRIA3
|
glutamate receptor, ionotropic, AMPA 3 |
| chr22_+_39101728 | 0.85 |
ENST00000216044.5
ENST00000484657.1 |
GTPBP1
|
GTP binding protein 1 |
| chr19_-_47287990 | 0.83 |
ENST00000593713.1
ENST00000598022.1 ENST00000434726.2 |
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr14_+_74058410 | 0.83 |
ENST00000326303.4
|
ACOT4
|
acyl-CoA thioesterase 4 |
| chr3_-_191000172 | 0.82 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr3_-_164914640 | 0.80 |
ENST00000241274.3
|
SLITRK3
|
SLIT and NTRK-like family, member 3 |
| chr17_-_79876010 | 0.79 |
ENST00000328666.6
|
SIRT7
|
sirtuin 7 |
| chr15_-_88799661 | 0.79 |
ENST00000360948.2
ENST00000357724.2 ENST00000355254.2 ENST00000317501.3 |
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr11_+_60467047 | 0.77 |
ENST00000300226.2
|
MS4A8
|
membrane-spanning 4-domains, subfamily A, member 8 |
| chr10_-_4285923 | 0.76 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr1_+_50569575 | 0.72 |
ENST00000371827.1
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr3_-_46735132 | 0.70 |
ENST00000415953.1
|
ALS2CL
|
ALS2 C-terminal like |
| chr3_-_24207039 | 0.70 |
ENST00000280696.5
|
THRB
|
thyroid hormone receptor, beta |
| chr14_-_74892805 | 0.69 |
ENST00000331628.3
ENST00000554953.1 |
SYNDIG1L
|
synapse differentiation inducing 1-like |
| chr16_+_31044413 | 0.68 |
ENST00000394998.1
|
STX4
|
syntaxin 4 |
| chr3_+_32859510 | 0.68 |
ENST00000383763.5
|
TRIM71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr2_-_197457335 | 0.67 |
ENST00000260983.3
|
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr12_-_21487829 | 0.67 |
ENST00000445053.1
ENST00000452078.1 ENST00000458504.1 ENST00000422327.1 ENST00000421294.1 |
SLCO1A2
|
solute carrier organic anion transporter family, member 1A2 |
| chr22_+_51176624 | 0.66 |
ENST00000216139.5
ENST00000529621.1 |
ACR
|
acrosin |
| chr1_+_212738676 | 0.66 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr5_-_20575959 | 0.65 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr1_+_10292308 | 0.63 |
ENST00000377081.1
|
KIF1B
|
kinesin family member 1B |
| chr14_-_27066636 | 0.62 |
ENST00000267422.7
ENST00000344429.5 ENST00000574031.1 ENST00000465357.2 ENST00000547619.1 |
NOVA1
|
neuro-oncological ventral antigen 1 |
| chr11_-_5531215 | 0.60 |
ENST00000311659.4
|
UBQLN3
|
ubiquilin 3 |
| chr1_-_26633067 | 0.60 |
ENST00000421827.2
ENST00000374215.1 ENST00000374223.1 ENST00000357089.4 ENST00000535108.1 ENST00000314675.7 ENST00000436301.2 ENST00000423664.1 ENST00000374221.3 |
UBXN11
|
UBX domain protein 11 |
| chr9_-_2844058 | 0.59 |
ENST00000397885.2
|
KIAA0020
|
KIAA0020 |
| chr12_-_95941987 | 0.59 |
ENST00000537435.2
|
USP44
|
ubiquitin specific peptidase 44 |
| chr17_+_41052808 | 0.59 |
ENST00000592383.1
ENST00000253801.2 ENST00000585489.1 |
G6PC
|
glucose-6-phosphatase, catalytic subunit |
| chr19_+_45449228 | 0.58 |
ENST00000252490.4
|
APOC2
|
apolipoprotein C-II |
| chr19_+_45449266 | 0.55 |
ENST00000592257.1
|
APOC2
|
apolipoprotein C-II |
| chr3_+_98699880 | 0.55 |
ENST00000473756.1
|
LINC00973
|
long intergenic non-protein coding RNA 973 |
| chr20_+_36373032 | 0.55 |
ENST00000373473.1
|
CTNNBL1
|
catenin, beta like 1 |
| chr2_+_27255806 | 0.54 |
ENST00000238788.9
ENST00000404032.3 |
TMEM214
|
transmembrane protein 214 |
| chr19_+_46800289 | 0.54 |
ENST00000377670.4
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit |
| chr17_+_56232494 | 0.53 |
ENST00000268912.5
|
OR4D1
|
olfactory receptor, family 4, subfamily D, member 1 |
| chr10_+_97759848 | 0.53 |
ENST00000424464.1
ENST00000410012.2 ENST00000344386.3 |
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chr18_-_49557 | 0.53 |
ENST00000308911.6
|
RP11-683L23.1
|
Tubulin beta-8 chain-like protein LOC260334 |
| chr12_+_21679220 | 0.52 |
ENST00000256969.2
|
C12orf39
|
chromosome 12 open reading frame 39 |
| chr4_+_107237660 | 0.51 |
ENST00000394701.4
|
AIMP1
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 |
| chr3_-_46735155 | 0.50 |
ENST00000318962.4
|
ALS2CL
|
ALS2 C-terminal like |
| chr2_-_161350305 | 0.50 |
ENST00000348849.3
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1 |
| chr11_-_126870655 | 0.49 |
ENST00000525144.2
|
KIRREL3
|
kin of IRRE like 3 (Drosophila) |
| chr10_-_46167722 | 0.49 |
ENST00000374366.3
ENST00000344646.5 |
ZFAND4
|
zinc finger, AN1-type domain 4 |
| chr7_+_141463897 | 0.48 |
ENST00000247879.2
|
TAS2R3
|
taste receptor, type 2, member 3 |
| chr8_-_23712312 | 0.47 |
ENST00000290271.2
|
STC1
|
stanniocalcin 1 |
| chr4_-_111563076 | 0.47 |
ENST00000354925.2
ENST00000511990.1 |
PITX2
|
paired-like homeodomain 2 |
| chr5_-_75008244 | 0.46 |
ENST00000510798.1
ENST00000446329.2 |
POC5
|
POC5 centriolar protein |
| chr3_+_181429704 | 0.46 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr10_-_102046098 | 0.44 |
ENST00000441611.1
|
BLOC1S2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr10_-_102046417 | 0.44 |
ENST00000370372.2
|
BLOC1S2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr1_-_177939348 | 0.43 |
ENST00000464631.2
|
SEC16B
|
SEC16 homolog B (S. cerevisiae) |
| chr15_+_31196080 | 0.43 |
ENST00000561607.1
ENST00000565466.1 |
FAN1
|
FANCD2/FANCI-associated nuclease 1 |
| chr5_-_115910630 | 0.43 |
ENST00000343348.6
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr11_+_118938485 | 0.42 |
ENST00000300793.6
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae) |
| chr5_-_60458179 | 0.42 |
ENST00000507416.1
ENST00000339020.3 |
SMIM15
|
small integral membrane protein 15 |
| chr5_-_171881491 | 0.42 |
ENST00000311601.5
|
SH3PXD2B
|
SH3 and PX domains 2B |
| chr13_+_30002741 | 0.42 |
ENST00000380808.2
|
MTUS2
|
microtubule associated tumor suppressor candidate 2 |
| chr17_-_39216344 | 0.41 |
ENST00000391418.2
|
KRTAP2-3
|
keratin associated protein 2-3 |
| chr12_-_11422739 | 0.41 |
ENST00000279573.7
|
PRB3
|
proline-rich protein BstNI subfamily 3 |
| chr11_+_4788500 | 0.40 |
ENST00000380390.1
|
MMP26
|
matrix metallopeptidase 26 |
| chr3_+_108308513 | 0.40 |
ENST00000361582.3
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr7_+_97736197 | 0.40 |
ENST00000297293.5
|
LMTK2
|
lemur tyrosine kinase 2 |
| chr7_+_148844516 | 0.39 |
ENST00000420008.2
ENST00000475153.1 |
ZNF398
|
zinc finger protein 398 |
| chr16_+_27214802 | 0.39 |
ENST00000380948.2
ENST00000286096.4 |
KDM8
|
lysine (K)-specific demethylase 8 |
| chr9_+_130565487 | 0.39 |
ENST00000373225.3
ENST00000431857.1 |
FPGS
|
folylpolyglutamate synthase |
| chr10_-_94003003 | 0.38 |
ENST00000412050.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr2_+_89986318 | 0.37 |
ENST00000491977.1
|
IGKV2D-29
|
immunoglobulin kappa variable 2D-29 |
| chr1_+_26496362 | 0.37 |
ENST00000374266.5
ENST00000270812.5 |
ZNF593
|
zinc finger protein 593 |
| chr10_-_95209 | 0.37 |
ENST00000332708.5
ENST00000309812.4 |
TUBB8
|
tubulin, beta 8 class VIII |
| chr18_+_11490077 | 0.37 |
ENST00000586947.1
ENST00000591431.1 |
RP11-712C7.1
|
RP11-712C7.1 |
| chr1_+_50571949 | 0.36 |
ENST00000357083.4
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr1_-_116383322 | 0.36 |
ENST00000429731.1
|
NHLH2
|
nescient helix loop helix 2 |
| chr1_+_25598989 | 0.36 |
ENST00000454452.2
|
RHD
|
Rh blood group, D antigen |
| chr6_+_27925019 | 0.35 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6 |
| chr12_-_11422630 | 0.34 |
ENST00000381842.3
ENST00000538488.1 |
PRB3
|
proline-rich protein BstNI subfamily 3 |
| chr6_+_29079668 | 0.34 |
ENST00000377169.1
|
OR2J3
|
olfactory receptor, family 2, subfamily J, member 3 |
| chr7_-_152373216 | 0.34 |
ENST00000359321.1
|
XRCC2
|
X-ray repair complementing defective repair in Chinese hamster cells 2 |
| chrX_-_70326455 | 0.34 |
ENST00000374251.5
|
CXorf65
|
chromosome X open reading frame 65 |
| chr7_-_6523688 | 0.34 |
ENST00000490996.1
|
KDELR2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr13_+_52598827 | 0.33 |
ENST00000521776.2
|
UTP14C
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog C (yeast) |
| chr1_+_78245303 | 0.33 |
ENST00000370791.3
ENST00000443751.2 |
FAM73A
|
family with sequence similarity 73, member A |
| chr22_-_37505588 | 0.33 |
ENST00000406856.1
|
TMPRSS6
|
transmembrane protease, serine 6 |
| chr6_+_116782527 | 0.33 |
ENST00000368606.3
ENST00000368605.1 |
FAM26F
|
family with sequence similarity 26, member F |
| chr16_-_69788816 | 0.33 |
ENST00000268802.5
|
NOB1
|
NIN1/RPN12 binding protein 1 homolog (S. cerevisiae) |
| chr5_+_169780485 | 0.32 |
ENST00000377360.4
|
KCNIP1
|
Kv channel interacting protein 1 |
| chr17_-_64225508 | 0.32 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr2_-_214016314 | 0.32 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr3_-_52273098 | 0.32 |
ENST00000499914.2
ENST00000305533.5 ENST00000597542.1 |
TWF2
TLR9
|
twinfilin actin-binding protein 2 toll-like receptor 9 |
| chr1_-_32384693 | 0.31 |
ENST00000602683.1
ENST00000470404.1 |
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2 |
| chr1_-_116383738 | 0.31 |
ENST00000320238.3
|
NHLH2
|
nescient helix loop helix 2 |
| chr22_-_37505449 | 0.31 |
ENST00000406725.1
|
TMPRSS6
|
transmembrane protease, serine 6 |
| chr1_+_86934526 | 0.31 |
ENST00000394711.1
|
CLCA1
|
chloride channel accessory 1 |
| chr19_-_49243845 | 0.30 |
ENST00000222145.4
|
RASIP1
|
Ras interacting protein 1 |
| chr19_+_45596398 | 0.30 |
ENST00000544069.2
|
PPP1R37
|
protein phosphatase 1, regulatory subunit 37 |
| chr13_-_36920420 | 0.30 |
ENST00000438666.2
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr16_-_75467318 | 0.30 |
ENST00000283882.3
|
CFDP1
|
craniofacial development protein 1 |
| chr5_+_42756903 | 0.30 |
ENST00000361970.5
ENST00000388827.4 |
CCDC152
|
coiled-coil domain containing 152 |
| chr10_+_98064085 | 0.29 |
ENST00000419175.1
ENST00000371174.2 |
DNTT
|
DNA nucleotidylexotransferase |
| chr1_+_113933371 | 0.29 |
ENST00000369617.4
|
MAGI3
|
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
| chr11_-_47788847 | 0.29 |
ENST00000263773.5
|
FNBP4
|
formin binding protein 4 |
| chr22_-_32146106 | 0.29 |
ENST00000327423.6
ENST00000397493.2 ENST00000434485.1 ENST00000412743.1 |
PRR14L
|
proline rich 14-like |
| chrX_+_70435044 | 0.29 |
ENST00000374029.1
ENST00000374022.3 ENST00000447581.1 |
GJB1
|
gap junction protein, beta 1, 32kDa |
| chr18_-_31802282 | 0.28 |
ENST00000535475.1
|
NOL4
|
nucleolar protein 4 |
| chr3_-_170744498 | 0.28 |
ENST00000382808.4
ENST00000314251.3 |
SLC2A2
|
solute carrier family 2 (facilitated glucose transporter), member 2 |
| chr2_-_172967621 | 0.28 |
ENST00000234198.4
ENST00000466293.2 |
DLX2
|
distal-less homeobox 2 |
| chr1_-_202679535 | 0.27 |
ENST00000367268.4
|
SYT2
|
synaptotagmin II |
| chrX_+_49126294 | 0.27 |
ENST00000466508.1
ENST00000438316.1 ENST00000055335.6 ENST00000495799.1 |
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F |
| chr1_+_156589051 | 0.27 |
ENST00000255039.1
|
HAPLN2
|
hyaluronan and proteoglycan link protein 2 |
| chr4_+_169418195 | 0.26 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr2_-_161349909 | 0.26 |
ENST00000392753.3
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1 |
| chr1_-_197036364 | 0.26 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr14_+_37131058 | 0.26 |
ENST00000361487.6
|
PAX9
|
paired box 9 |
| chr2_-_39348137 | 0.25 |
ENST00000426016.1
|
SOS1
|
son of sevenless homolog 1 (Drosophila) |
| chr2_+_241375069 | 0.25 |
ENST00000264039.2
|
GPC1
|
glypican 1 |
| chr17_-_56082455 | 0.25 |
ENST00000578794.1
|
RP11-159D12.5
|
Uncharacterized protein |
| chr1_-_159684371 | 0.24 |
ENST00000255030.5
ENST00000437342.1 ENST00000368112.1 ENST00000368111.1 ENST00000368110.1 ENST00000343919.2 |
CRP
|
C-reactive protein, pentraxin-related |
| chr8_-_70983506 | 0.24 |
ENST00000276594.2
|
PRDM14
|
PR domain containing 14 |
| chr15_-_38519066 | 0.24 |
ENST00000561320.1
ENST00000561161.1 |
RP11-346D14.1
|
RP11-346D14.1 |
| chr8_+_120885949 | 0.24 |
ENST00000523492.1
ENST00000286234.5 |
DEPTOR
|
DEP domain containing MTOR-interacting protein |
| chr2_+_108602971 | 0.24 |
ENST00000409059.1
ENST00000540517.1 ENST00000264047.2 |
SLC5A7
|
solute carrier family 5 (sodium/choline cotransporter), member 7 |
| chr19_+_45596218 | 0.24 |
ENST00000421905.1
ENST00000221462.4 |
PPP1R37
|
protein phosphatase 1, regulatory subunit 37 |
| chr9_+_140135665 | 0.23 |
ENST00000340384.4
|
TUBB4B
|
tubulin, beta 4B class IVb |
| chrX_+_135388147 | 0.23 |
ENST00000394141.1
|
GPR112
|
G protein-coupled receptor 112 |
| chr4_+_71091786 | 0.22 |
ENST00000317987.5
|
FDCSP
|
follicular dendritic cell secreted protein |
| chr15_-_99789736 | 0.22 |
ENST00000560235.1
ENST00000394132.2 ENST00000560860.1 ENST00000558078.1 ENST00000394136.1 ENST00000262074.4 ENST00000558613.1 ENST00000394130.1 ENST00000560772.1 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr3_+_186383741 | 0.22 |
ENST00000232003.4
|
HRG
|
histidine-rich glycoprotein |
| chr9_+_95726243 | 0.22 |
ENST00000416701.2
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chrX_-_151143140 | 0.22 |
ENST00000393914.3
ENST00000370328.3 ENST00000370325.1 |
GABRE
|
gamma-aminobutyric acid (GABA) A receptor, epsilon |
| chr19_+_2785458 | 0.21 |
ENST00000307741.6
ENST00000585338.1 |
THOP1
|
thimet oligopeptidase 1 |
| chr3_+_148415571 | 0.21 |
ENST00000497524.1
ENST00000349243.3 ENST00000542281.1 ENST00000418473.2 ENST00000404754.2 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr18_+_49866496 | 0.21 |
ENST00000442544.2
|
DCC
|
deleted in colorectal carcinoma |
| chrX_-_110654147 | 0.21 |
ENST00000358070.4
|
DCX
|
doublecortin |
| chr5_-_151784838 | 0.21 |
ENST00000255262.3
|
NMUR2
|
neuromedin U receptor 2 |
| chr4_-_103747011 | 0.20 |
ENST00000350435.7
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr10_+_51576285 | 0.19 |
ENST00000443446.1
|
NCOA4
|
nuclear receptor coactivator 4 |
| chrX_+_122318006 | 0.19 |
ENST00000371266.1
ENST00000264357.5 |
GRIA3
|
glutamate receptor, ionotropic, AMPA 3 |
| chr8_-_33370607 | 0.19 |
ENST00000360742.5
ENST00000523305.1 |
TTI2
|
TELO2 interacting protein 2 |
| chr18_-_31802056 | 0.18 |
ENST00000538587.1
|
NOL4
|
nucleolar protein 4 |
| chr12_+_15475462 | 0.18 |
ENST00000543886.1
ENST00000348962.2 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr11_+_60995849 | 0.17 |
ENST00000537932.1
|
PGA4
|
pepsinogen 4, group I (pepsinogen A) |
| chr1_+_57320437 | 0.17 |
ENST00000361249.3
|
C8A
|
complement component 8, alpha polypeptide |
| chr2_+_234959376 | 0.17 |
ENST00000425558.1
|
SPP2
|
secreted phosphoprotein 2, 24kDa |
| chr6_-_46048116 | 0.17 |
ENST00000185206.6
|
CLIC5
|
chloride intracellular channel 5 |
| chr7_-_154794763 | 0.17 |
ENST00000404141.1
|
PAXIP1
|
PAX interacting (with transcription-activation domain) protein 1 |
| chr11_-_10828892 | 0.17 |
ENST00000525681.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr3_+_188889737 | 0.16 |
ENST00000345063.3
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr1_+_156756667 | 0.15 |
ENST00000526188.1
ENST00000454659.1 |
PRCC
|
papillary renal cell carcinoma (translocation-associated) |
| chr1_+_145301735 | 0.15 |
ENST00000605176.1
|
NBPF10
|
neuroblastoma breakpoint family, member 10 |
| chr19_-_46916805 | 0.15 |
ENST00000307522.3
|
CCDC8
|
coiled-coil domain containing 8 |
| chr5_-_135701164 | 0.15 |
ENST00000355180.3
ENST00000426057.2 ENST00000513104.1 |
TRPC7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chrX_+_14547632 | 0.14 |
ENST00000218075.4
|
GLRA2
|
glycine receptor, alpha 2 |
| chrX_-_131262048 | 0.14 |
ENST00000298542.4
|
FRMD7
|
FERM domain containing 7 |
| chr11_-_95657231 | 0.14 |
ENST00000409459.1
ENST00000352297.7 ENST00000393223.3 ENST00000346299.5 |
MTMR2
|
myotubularin related protein 2 |
| chr13_-_86373536 | 0.14 |
ENST00000400286.2
|
SLITRK6
|
SLIT and NTRK-like family, member 6 |
| chr3_-_112693759 | 0.14 |
ENST00000440122.2
ENST00000490004.1 |
CD200R1
|
CD200 receptor 1 |
| chr17_-_50235423 | 0.14 |
ENST00000340813.6
|
CA10
|
carbonic anhydrase X |
| chr20_+_23471727 | 0.14 |
ENST00000449810.1
ENST00000246012.1 |
CST8
|
cystatin 8 (cystatin-related epididymal specific) |
| chr8_-_101719159 | 0.13 |
ENST00000520868.1
ENST00000522658.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr6_-_133055815 | 0.13 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr1_+_17575584 | 0.13 |
ENST00000375460.3
|
PADI3
|
peptidyl arginine deiminase, type III |
Network of associatons between targets according to the STRING database.
First level regulatory network of ONECUT1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.4 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 1.0 | 3.1 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.6 | 1.8 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.6 | 2.4 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.5 | 2.0 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.4 | 4.6 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.4 | 1.8 | GO:0060356 | leucine import(GO:0060356) |
| 0.3 | 0.8 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.3 | 0.8 | GO:0048687 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.2 | 3.0 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.2 | 0.9 | GO:0019605 | butyrate metabolic process(GO:0019605) |
| 0.2 | 0.7 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.2 | 2.1 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.2 | 0.7 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.2 | 0.7 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.2 | 0.8 | GO:0010585 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.2 | 1.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.2 | 1.0 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.2 | 0.9 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.2 | 0.6 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.2 | 0.5 | GO:0021763 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.1 | 3.8 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 1.6 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.1 | 0.4 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 0.4 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 0.7 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 0.3 | GO:0021893 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.1 | 1.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 0.3 | GO:1901895 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.1 | 1.9 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.1 | 0.3 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 0.4 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.1 | 0.2 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.1 | 0.3 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 0.2 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.1 | 0.2 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.1 | 0.9 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.1 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.2 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.1 | 0.3 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.1 | 0.6 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.2 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 0.5 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.2 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.2 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.5 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.0 | 1.0 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.5 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.6 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.4 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.7 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.4 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 1.1 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.3 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 1.1 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.3 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 1.1 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.6 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.3 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.2 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.4 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.7 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.6 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.4 | GO:0070863 | peroxisome fission(GO:0016559) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.1 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 1.1 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 1.0 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.2 | GO:0032930 | positive regulation of superoxide anion generation(GO:0032930) |
| 0.0 | 0.4 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 0.2 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.9 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.2 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.3 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 | 3.4 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.3 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 1.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.7 | GO:0030071 | regulation of mitotic metaphase/anaphase transition(GO:0030071) |
| 0.0 | 0.5 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.3 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 1.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 0.7 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.2 | 0.7 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 1.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.5 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.1 | 2.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.0 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.7 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 0.4 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.8 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.3 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.9 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.5 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 3.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.3 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.4 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 1.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 1.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 3.9 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.2 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 1.8 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 7.3 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.6 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 2.4 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.0 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 2.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) organelle outer membrane(GO:0031968) |
| 0.0 | 1.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.4 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.4 | 1.8 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.4 | 3.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.4 | 2.7 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.3 | 0.8 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.2 | 1.9 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 0.7 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.2 | 0.5 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.2 | 0.7 | GO:0004040 | amidase activity(GO:0004040) |
| 0.1 | 0.6 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 3.9 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.4 | GO:0031177 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.8 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 1.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.5 | GO:0033265 | choline binding(GO:0033265) |
| 0.1 | 3.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.3 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.1 | 0.3 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.1 | 0.9 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.3 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 0.7 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 1.0 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.8 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 1.1 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.8 | GO:0005030 | GPI-linked ephrin receptor activity(GO:0005004) neurotrophin receptor activity(GO:0005030) |
| 0.1 | 2.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 1.1 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.9 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 1.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.3 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.7 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.0 | 0.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 1.1 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 1.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.1 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.2 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 5.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 2.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 3.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 1.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 3.6 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 2.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 3.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.1 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.9 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 3.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 2.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 4.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 4.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.0 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.0 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 3.7 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 1.2 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 2.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.9 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.8 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.7 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.6 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |