Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
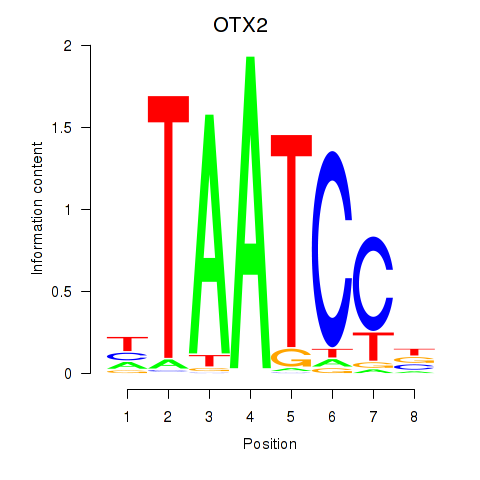
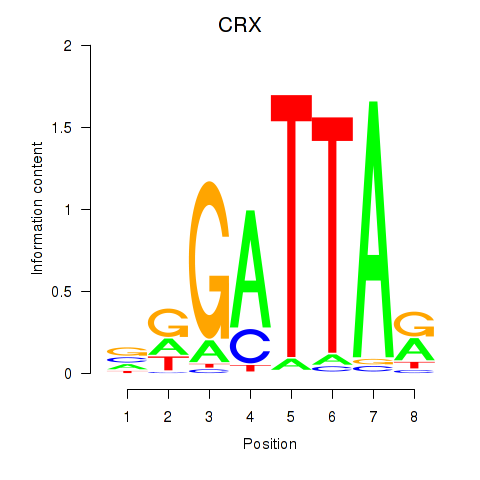
Results for OTX2_CRX
Z-value: 0.46
Transcription factors associated with OTX2_CRX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OTX2
|
ENSG00000165588.12 | orthodenticle homeobox 2 |
|
CRX
|
ENSG00000105392.11 | cone-rod homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| OTX2 | hg19_v2_chr14_-_57272366_57272391 | -0.34 | 1.0e-01 | Click! |
| CRX | hg19_v2_chr19_+_48325097_48325211 | 0.34 | 1.0e-01 | Click! |
Activity profile of OTX2_CRX motif
Sorted Z-values of OTX2_CRX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_6949964 | 1.63 |
ENST00000541978.1
ENST00000435982.2 |
GNB3
|
guanine nucleotide binding protein (G protein), beta polypeptide 3 |
| chr6_+_130339710 | 0.76 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr14_-_95236551 | 0.61 |
ENST00000238558.3
|
GSC
|
goosecoid homeobox |
| chr1_-_165414414 | 0.42 |
ENST00000359842.5
|
RXRG
|
retinoid X receptor, gamma |
| chr11_-_62752162 | 0.40 |
ENST00000458333.2
ENST00000421062.2 |
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr19_+_46001697 | 0.39 |
ENST00000451287.2
ENST00000324688.4 |
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr12_+_15125954 | 0.36 |
ENST00000266395.2
|
PDE6H
|
phosphodiesterase 6H, cGMP-specific, cone, gamma |
| chr11_+_61248583 | 0.35 |
ENST00000432063.2
ENST00000338608.2 |
PPP1R32
|
protein phosphatase 1, regulatory subunit 32 |
| chr19_+_2476116 | 0.35 |
ENST00000215631.4
ENST00000587345.1 |
GADD45B
|
growth arrest and DNA-damage-inducible, beta |
| chr12_+_85673868 | 0.34 |
ENST00000316824.3
|
ALX1
|
ALX homeobox 1 |
| chr6_-_42690312 | 0.34 |
ENST00000230381.5
|
PRPH2
|
peripherin 2 (retinal degeneration, slow) |
| chr10_+_103986085 | 0.29 |
ENST00000370005.3
|
ELOVL3
|
ELOVL fatty acid elongase 3 |
| chr14_-_22005018 | 0.28 |
ENST00000546363.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr19_+_48325097 | 0.28 |
ENST00000221996.7
ENST00000539067.1 |
CRX
|
cone-rod homeobox |
| chr22_-_38380543 | 0.26 |
ENST00000396884.2
|
SOX10
|
SRY (sex determining region Y)-box 10 |
| chr8_+_54764346 | 0.25 |
ENST00000297313.3
ENST00000344277.6 |
RGS20
|
regulator of G-protein signaling 20 |
| chr13_-_33780133 | 0.25 |
ENST00000399365.3
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr11_+_67222818 | 0.24 |
ENST00000325656.5
|
CABP4
|
calcium binding protein 4 |
| chr14_-_22005062 | 0.23 |
ENST00000317492.5
|
SALL2
|
spalt-like transcription factor 2 |
| chr5_-_88179302 | 0.23 |
ENST00000504921.2
|
MEF2C
|
myocyte enhancer factor 2C |
| chr4_+_129730947 | 0.22 |
ENST00000452328.2
ENST00000504089.1 |
PHF17
|
jade family PHD finger 1 |
| chr8_-_133123406 | 0.22 |
ENST00000434736.2
|
HHLA1
|
HERV-H LTR-associating 1 |
| chr17_-_40264692 | 0.22 |
ENST00000591220.1
ENST00000251642.3 |
DHX58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr2_+_66662510 | 0.22 |
ENST00000272369.9
ENST00000407092.2 |
MEIS1
|
Meis homeobox 1 |
| chr20_-_18774614 | 0.21 |
ENST00000412553.1
|
LINC00652
|
long intergenic non-protein coding RNA 652 |
| chr3_-_54962100 | 0.21 |
ENST00000273286.5
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1 |
| chr2_+_66662690 | 0.21 |
ENST00000488550.1
|
MEIS1
|
Meis homeobox 1 |
| chr4_+_129730779 | 0.21 |
ENST00000226319.6
|
PHF17
|
jade family PHD finger 1 |
| chr1_+_144989309 | 0.21 |
ENST00000596396.1
|
AL590452.1
|
Uncharacterized protein |
| chr1_+_162351503 | 0.20 |
ENST00000458626.2
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr6_+_35310312 | 0.20 |
ENST00000448077.2
ENST00000360694.3 ENST00000418635.2 ENST00000444397.1 |
PPARD
|
peroxisome proliferator-activated receptor delta |
| chr1_+_244214577 | 0.19 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr3_-_112218205 | 0.18 |
ENST00000383680.4
|
BTLA
|
B and T lymphocyte associated |
| chr3_-_50649192 | 0.18 |
ENST00000443053.2
ENST00000348721.3 |
CISH
|
cytokine inducible SH2-containing protein |
| chr3_+_63953415 | 0.18 |
ENST00000484332.1
|
ATXN7
|
ataxin 7 |
| chr6_-_29399744 | 0.17 |
ENST00000377154.1
|
OR5V1
|
olfactory receptor, family 5, subfamily V, member 1 |
| chr2_-_89459813 | 0.17 |
ENST00000390256.2
|
IGKV6-21
|
immunoglobulin kappa variable 6-21 (non-functional) |
| chrX_-_21676442 | 0.17 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr11_-_118436606 | 0.17 |
ENST00000530872.1
|
IFT46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr7_-_100171270 | 0.17 |
ENST00000538735.1
|
SAP25
|
Sin3A-associated protein, 25kDa |
| chr15_+_41221536 | 0.17 |
ENST00000249749.5
|
DLL4
|
delta-like 4 (Drosophila) |
| chrX_+_15767971 | 0.16 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr7_+_73868220 | 0.16 |
ENST00000455841.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr14_-_22005343 | 0.16 |
ENST00000327430.3
|
SALL2
|
spalt-like transcription factor 2 |
| chr10_-_126716459 | 0.16 |
ENST00000309035.6
|
CTBP2
|
C-terminal binding protein 2 |
| chr13_-_95131923 | 0.16 |
ENST00000377028.5
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr18_+_32290218 | 0.16 |
ENST00000348997.5
ENST00000588949.1 ENST00000597599.1 |
DTNA
|
dystrobrevin, alpha |
| chr12_+_81101277 | 0.16 |
ENST00000228641.3
|
MYF6
|
myogenic factor 6 (herculin) |
| chrX_+_129473859 | 0.15 |
ENST00000424447.1
|
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr6_+_34433844 | 0.15 |
ENST00000244458.2
ENST00000374043.2 |
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1 |
| chr3_-_190580404 | 0.15 |
ENST00000442080.1
|
GMNC
|
geminin coiled-coil domain containing |
| chr17_-_46690839 | 0.15 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr11_-_118436707 | 0.15 |
ENST00000264020.2
ENST00000264021.3 |
IFT46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chrX_-_124097620 | 0.14 |
ENST00000371130.3
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr11_-_128712362 | 0.14 |
ENST00000392664.2
|
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr1_-_117664317 | 0.14 |
ENST00000256649.4
ENST00000369464.3 ENST00000485032.1 |
TRIM45
|
tripartite motif containing 45 |
| chr17_+_68100989 | 0.14 |
ENST00000585558.1
ENST00000392670.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr15_+_96869165 | 0.14 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chrX_+_17653413 | 0.14 |
ENST00000398097.3
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
| chr16_-_31076273 | 0.13 |
ENST00000426488.2
|
ZNF668
|
zinc finger protein 668 |
| chr5_+_5140436 | 0.13 |
ENST00000511368.1
ENST00000274181.7 |
ADAMTS16
|
ADAM metallopeptidase with thrombospondin type 1 motif, 16 |
| chr14_+_88471468 | 0.13 |
ENST00000267549.3
|
GPR65
|
G protein-coupled receptor 65 |
| chr3_-_112218378 | 0.13 |
ENST00000334529.5
|
BTLA
|
B and T lymphocyte associated |
| chr4_-_153601136 | 0.13 |
ENST00000504064.1
ENST00000304385.3 |
TMEM154
|
transmembrane protein 154 |
| chr2_+_170366203 | 0.13 |
ENST00000284669.1
|
KLHL41
|
kelch-like family member 41 |
| chr11_-_130184555 | 0.13 |
ENST00000525842.1
|
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr2_-_121223697 | 0.13 |
ENST00000593290.1
|
FLJ14816
|
long intergenic non-protein coding RNA 1101 |
| chr6_-_170599561 | 0.12 |
ENST00000366756.3
|
DLL1
|
delta-like 1 (Drosophila) |
| chr3_-_49058479 | 0.12 |
ENST00000440857.1
|
DALRD3
|
DALR anticodon binding domain containing 3 |
| chr18_-_64271363 | 0.12 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr2_+_33661382 | 0.12 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr16_-_31076332 | 0.12 |
ENST00000539836.3
ENST00000535577.1 ENST00000442862.2 |
ZNF668
|
zinc finger protein 668 |
| chr12_+_53693466 | 0.12 |
ENST00000267103.5
ENST00000548632.1 |
C12orf10
|
chromosome 12 open reading frame 10 |
| chr13_-_52027134 | 0.12 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr5_+_161494770 | 0.12 |
ENST00000414552.2
ENST00000361925.4 |
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr9_-_39288092 | 0.12 |
ENST00000323947.7
ENST00000297668.6 ENST00000377656.2 ENST00000377659.1 |
CNTNAP3
|
contactin associated protein-like 3 |
| chr1_+_207277590 | 0.12 |
ENST00000367070.3
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr17_+_7905912 | 0.12 |
ENST00000254854.4
|
GUCY2D
|
guanylate cyclase 2D, membrane (retina-specific) |
| chr5_-_524445 | 0.12 |
ENST00000514375.1
ENST00000264938.3 |
SLC9A3
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 |
| chr20_+_59654146 | 0.12 |
ENST00000441660.1
|
RP5-827L5.1
|
RP5-827L5.1 |
| chr12_+_53693812 | 0.11 |
ENST00000549488.1
|
C12orf10
|
chromosome 12 open reading frame 10 |
| chrX_-_10588459 | 0.11 |
ENST00000380782.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr5_+_63461642 | 0.11 |
ENST00000296615.6
ENST00000381081.2 ENST00000389100.4 |
RNF180
|
ring finger protein 180 |
| chr5_-_88179017 | 0.11 |
ENST00000514028.1
ENST00000514015.1 ENST00000503075.1 ENST00000437473.2 |
MEF2C
|
myocyte enhancer factor 2C |
| chr2_+_201170703 | 0.11 |
ENST00000358677.5
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr2_-_188312971 | 0.11 |
ENST00000410068.1
ENST00000447403.1 ENST00000410102.1 |
CALCRL
|
calcitonin receptor-like |
| chr17_-_38859996 | 0.11 |
ENST00000264651.2
|
KRT24
|
keratin 24 |
| chr5_-_88178964 | 0.11 |
ENST00000513252.1
ENST00000508569.1 ENST00000510942.1 ENST00000506554.1 |
MEF2C
|
myocyte enhancer factor 2C |
| chr6_+_35310391 | 0.11 |
ENST00000337400.2
ENST00000311565.4 ENST00000540939.1 |
PPARD
|
peroxisome proliferator-activated receptor delta |
| chrX_-_10588595 | 0.10 |
ENST00000423614.1
ENST00000317552.4 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr12_-_53189892 | 0.10 |
ENST00000309505.3
ENST00000417996.2 |
KRT3
|
keratin 3 |
| chr17_+_4643337 | 0.10 |
ENST00000592813.1
|
ZMYND15
|
zinc finger, MYND-type containing 15 |
| chr2_-_70780572 | 0.10 |
ENST00000450929.1
|
TGFA
|
transforming growth factor, alpha |
| chr7_+_90338712 | 0.10 |
ENST00000265741.3
ENST00000406263.1 |
CDK14
|
cyclin-dependent kinase 14 |
| chr1_+_55505184 | 0.10 |
ENST00000302118.5
|
PCSK9
|
proprotein convertase subtilisin/kexin type 9 |
| chr17_+_4643300 | 0.10 |
ENST00000433935.1
|
ZMYND15
|
zinc finger, MYND-type containing 15 |
| chr9_-_130966497 | 0.10 |
ENST00000393608.1
ENST00000372948.3 |
CIZ1
|
CDKN1A interacting zinc finger protein 1 |
| chr5_+_161494521 | 0.10 |
ENST00000356592.3
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr5_-_149669192 | 0.10 |
ENST00000398376.3
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr17_-_9808887 | 0.10 |
ENST00000226193.5
|
RCVRN
|
recoverin |
| chr1_+_11751748 | 0.09 |
ENST00000294485.5
|
DRAXIN
|
dorsal inhibitory axon guidance protein |
| chr6_-_111804905 | 0.09 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr2_-_14541060 | 0.09 |
ENST00000418420.1
ENST00000417751.1 |
LINC00276
|
long intergenic non-protein coding RNA 276 |
| chr15_+_58430368 | 0.09 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr3_-_137851220 | 0.09 |
ENST00000236709.3
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr10_+_18240834 | 0.09 |
ENST00000377371.3
ENST00000539911.1 |
SLC39A12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr4_+_154125565 | 0.09 |
ENST00000338700.5
|
TRIM2
|
tripartite motif containing 2 |
| chr19_-_48547294 | 0.09 |
ENST00000293255.2
|
CABP5
|
calcium binding protein 5 |
| chr9_-_139361503 | 0.09 |
ENST00000453963.1
|
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr14_-_36789865 | 0.09 |
ENST00000416007.4
|
MBIP
|
MAP3K12 binding inhibitory protein 1 |
| chr3_+_4535025 | 0.09 |
ENST00000302640.8
ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr14_-_36789783 | 0.09 |
ENST00000605579.1
ENST00000604336.1 ENST00000359527.7 ENST00000603139.1 ENST00000318473.7 |
MBIP
|
MAP3K12 binding inhibitory protein 1 |
| chr4_-_100484825 | 0.09 |
ENST00000273962.3
ENST00000514547.1 ENST00000455368.2 |
TRMT10A
|
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr15_+_58430567 | 0.08 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr14_-_67981870 | 0.08 |
ENST00000555994.1
|
TMEM229B
|
transmembrane protein 229B |
| chrM_+_9207 | 0.08 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chr4_-_100485143 | 0.08 |
ENST00000394877.3
|
TRMT10A
|
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr2_-_175499294 | 0.08 |
ENST00000392547.2
|
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr15_-_74495188 | 0.08 |
ENST00000563965.1
ENST00000395105.4 |
STRA6
|
stimulated by retinoic acid 6 |
| chr16_-_21663919 | 0.08 |
ENST00000569602.1
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr7_-_97881429 | 0.08 |
ENST00000420697.1
ENST00000379795.3 ENST00000415086.1 ENST00000542604.1 ENST00000447648.2 |
TECPR1
|
tectonin beta-propeller repeat containing 1 |
| chr15_-_65407524 | 0.08 |
ENST00000559089.1
|
UBAP1L
|
ubiquitin associated protein 1-like |
| chr12_-_53171128 | 0.08 |
ENST00000332411.2
|
KRT76
|
keratin 76 |
| chr18_+_616711 | 0.08 |
ENST00000579494.1
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr5_-_36301984 | 0.08 |
ENST00000502994.1
ENST00000515759.1 ENST00000296604.3 |
RANBP3L
|
RAN binding protein 3-like |
| chr11_-_130184470 | 0.07 |
ENST00000357899.4
ENST00000397753.1 |
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr12_-_118628350 | 0.07 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chrX_+_28605516 | 0.07 |
ENST00000378993.1
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1 |
| chr10_-_94003003 | 0.07 |
ENST00000412050.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr12_-_21487829 | 0.07 |
ENST00000445053.1
ENST00000452078.1 ENST00000458504.1 ENST00000422327.1 ENST00000421294.1 |
SLCO1A2
|
solute carrier organic anion transporter family, member 1A2 |
| chr1_+_66999268 | 0.07 |
ENST00000371039.1
ENST00000424320.1 |
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr19_+_11457175 | 0.07 |
ENST00000458408.1
ENST00000586451.1 ENST00000588592.1 |
CCDC159
|
coiled-coil domain containing 159 |
| chrX_-_119077695 | 0.07 |
ENST00000371410.3
|
NKAP
|
NFKB activating protein |
| chr7_+_73868439 | 0.07 |
ENST00000424337.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr1_-_102312517 | 0.07 |
ENST00000338858.5
|
OLFM3
|
olfactomedin 3 |
| chr12_-_54779511 | 0.07 |
ENST00000551109.1
ENST00000546970.1 |
ZNF385A
|
zinc finger protein 385A |
| chr5_+_173316341 | 0.07 |
ENST00000520867.1
ENST00000334035.5 |
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr11_-_67290867 | 0.07 |
ENST00000353903.5
ENST00000294288.4 |
CABP2
|
calcium binding protein 2 |
| chr8_+_70378852 | 0.07 |
ENST00000525061.1
ENST00000458141.2 ENST00000260128.4 |
SULF1
|
sulfatase 1 |
| chr9_-_20622478 | 0.07 |
ENST00000355930.6
ENST00000380338.4 |
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr10_+_18240753 | 0.07 |
ENST00000377369.2
|
SLC39A12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr3_-_47950745 | 0.07 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr12_-_90049878 | 0.07 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr2_+_192141611 | 0.06 |
ENST00000392316.1
|
MYO1B
|
myosin IB |
| chr18_-_64271316 | 0.06 |
ENST00000540086.1
ENST00000580157.1 |
CDH19
|
cadherin 19, type 2 |
| chr9_+_18474098 | 0.06 |
ENST00000327883.7
ENST00000431052.2 ENST00000380570.4 |
ADAMTSL1
|
ADAMTS-like 1 |
| chr18_+_616672 | 0.06 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr17_-_39661849 | 0.06 |
ENST00000246635.3
ENST00000336861.3 ENST00000587544.1 ENST00000587435.1 |
KRT13
|
keratin 13 |
| chr11_+_124735282 | 0.06 |
ENST00000397801.1
|
ROBO3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr2_+_120770686 | 0.06 |
ENST00000331393.4
ENST00000443124.1 |
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr10_-_105845674 | 0.06 |
ENST00000353479.5
ENST00000369733.3 |
COL17A1
|
collagen, type XVII, alpha 1 |
| chr17_-_48207157 | 0.06 |
ENST00000330175.4
ENST00000503131.1 |
SAMD14
|
sterile alpha motif domain containing 14 |
| chr12_+_133757995 | 0.06 |
ENST00000536435.2
ENST00000228289.5 ENST00000541211.2 ENST00000500625.3 ENST00000539248.2 ENST00000542711.2 ENST00000536899.2 ENST00000542986.2 ENST00000537565.1 ENST00000541975.2 |
ZNF268
|
zinc finger protein 268 |
| chr12_-_90049828 | 0.06 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr1_+_202431859 | 0.06 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr2_+_183943464 | 0.06 |
ENST00000354221.4
|
DUSP19
|
dual specificity phosphatase 19 |
| chr2_-_61765315 | 0.06 |
ENST00000406957.1
ENST00000401558.2 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr10_-_89577910 | 0.06 |
ENST00000308448.7
ENST00000541004.1 |
ATAD1
|
ATPase family, AAA domain containing 1 |
| chr2_+_201170596 | 0.06 |
ENST00000439084.1
ENST00000409718.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr7_-_104909435 | 0.05 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr12_-_56848426 | 0.05 |
ENST00000257979.4
|
MIP
|
major intrinsic protein of lens fiber |
| chr10_+_18240814 | 0.05 |
ENST00000377374.4
|
SLC39A12
|
solute carrier family 39 (zinc transporter), member 12 |
| chrX_+_41306575 | 0.05 |
ENST00000342595.2
ENST00000378220.1 |
NYX
|
nyctalopin |
| chr11_-_128894053 | 0.05 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr10_+_134150835 | 0.05 |
ENST00000432555.2
|
LRRC27
|
leucine rich repeat containing 27 |
| chr17_-_14683517 | 0.05 |
ENST00000379640.1
|
AC005863.1
|
AC005863.1 |
| chr11_+_36589547 | 0.05 |
ENST00000299440.5
|
RAG1
|
recombination activating gene 1 |
| chr5_-_150138061 | 0.05 |
ENST00000521533.1
ENST00000424236.1 |
DCTN4
|
dynactin 4 (p62) |
| chr3_-_186262166 | 0.05 |
ENST00000307944.5
|
CRYGS
|
crystallin, gamma S |
| chr2_-_55237484 | 0.05 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr15_+_33603147 | 0.05 |
ENST00000415757.3
ENST00000389232.4 |
RYR3
|
ryanodine receptor 3 |
| chr16_+_81272287 | 0.05 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr1_+_81771806 | 0.05 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr3_+_129247479 | 0.05 |
ENST00000296271.3
|
RHO
|
rhodopsin |
| chr2_+_219472637 | 0.05 |
ENST00000417849.1
|
PLCD4
|
phospholipase C, delta 4 |
| chr1_+_159141397 | 0.05 |
ENST00000368124.4
ENST00000368125.4 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr16_-_68269971 | 0.05 |
ENST00000565858.1
|
ESRP2
|
epithelial splicing regulatory protein 2 |
| chr19_+_18208603 | 0.05 |
ENST00000262811.6
|
MAST3
|
microtubule associated serine/threonine kinase 3 |
| chr1_+_167298281 | 0.05 |
ENST00000367862.5
|
POU2F1
|
POU class 2 homeobox 1 |
| chr12_-_55375622 | 0.05 |
ENST00000316577.8
|
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr2_+_120770581 | 0.05 |
ENST00000263713.5
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr12_+_7941989 | 0.05 |
ENST00000229307.4
|
NANOG
|
Nanog homeobox |
| chr19_-_42931567 | 0.05 |
ENST00000244289.4
|
LIPE
|
lipase, hormone-sensitive |
| chrX_-_153363188 | 0.05 |
ENST00000303391.6
|
MECP2
|
methyl CpG binding protein 2 (Rett syndrome) |
| chr7_+_73868120 | 0.05 |
ENST00000265755.3
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr11_-_30608413 | 0.05 |
ENST00000528686.1
|
MPPED2
|
metallophosphoesterase domain containing 2 |
| chr19_+_39989580 | 0.05 |
ENST00000596614.1
ENST00000205143.4 |
DLL3
|
delta-like 3 (Drosophila) |
| chr7_+_80231466 | 0.05 |
ENST00000309881.7
ENST00000534394.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr3_-_50605150 | 0.04 |
ENST00000357203.3
|
C3orf18
|
chromosome 3 open reading frame 18 |
| chr9_+_17134980 | 0.04 |
ENST00000380647.3
|
CNTLN
|
centlein, centrosomal protein |
| chrX_-_110655306 | 0.04 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr19_+_39989535 | 0.04 |
ENST00000356433.5
|
DLL3
|
delta-like 3 (Drosophila) |
| chr12_+_104697504 | 0.04 |
ENST00000527879.1
|
EID3
|
EP300 interacting inhibitor of differentiation 3 |
| chr17_-_43209862 | 0.04 |
ENST00000322765.5
|
PLCD3
|
phospholipase C, delta 3 |
| chr14_-_58894332 | 0.04 |
ENST00000395159.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr14_+_96722152 | 0.04 |
ENST00000216629.6
|
BDKRB1
|
bradykinin receptor B1 |
| chr9_-_95244781 | 0.04 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr10_-_62761188 | 0.04 |
ENST00000357917.4
|
RHOBTB1
|
Rho-related BTB domain containing 1 |
| chr2_-_225362533 | 0.04 |
ENST00000451538.1
|
CUL3
|
cullin 3 |
| chrM_+_8527 | 0.04 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of OTX2_CRX
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0014034 | neural crest cell fate commitment(GO:0014034) |
| 0.1 | 0.4 | GO:0097254 | renal tubular secretion(GO:0097254) |
| 0.1 | 0.1 | GO:0060844 | arterial endothelial cell fate commitment(GO:0060844) |
| 0.1 | 0.4 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.3 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.1 | 0.3 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.8 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.3 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.2 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.0 | 0.2 | GO:0072554 | blood vessel lumenization(GO:0072554) |
| 0.0 | 0.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.3 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.1 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.3 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 1.6 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.3 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.1 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.2 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.4 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.2 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.1 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.3 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.1 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.0 | 0.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.0 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.3 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.2 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.0 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.4 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.1 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.1 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:1903365 | regulation of fear response(GO:1903365) regulation of behavioral fear response(GO:2000822) |
| 0.0 | 0.0 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990667 | PCSK9-LDLR complex(GO:1990666) PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.7 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.2 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.2 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.0 | 0.2 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.2 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.1 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.0 | 0.3 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.4 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.2 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.2 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.0 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.0 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 0.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.0 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.4 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |