Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for PAX1_PAX9
Z-value: 0.49
Transcription factors associated with PAX1_PAX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
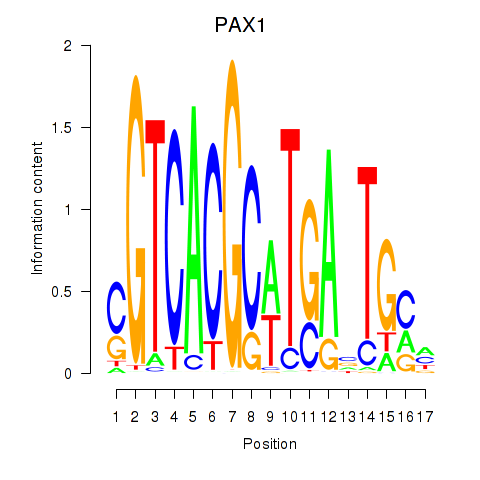
PAX1
|
ENSG00000125813.9 | paired box 1 |
|
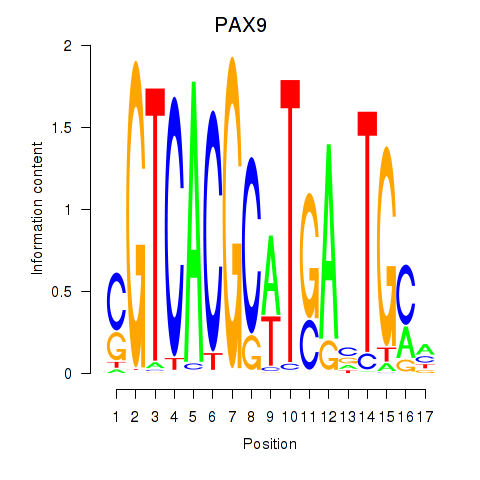
PAX9
|
ENSG00000198807.8 | paired box 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX9 | hg19_v2_chr14_+_37126765_37126799 | -0.14 | 5.0e-01 | Click! |
| PAX1 | hg19_v2_chr20_+_21686290_21686311 | -0.06 | 7.9e-01 | Click! |
Activity profile of PAX1_PAX9 motif
Sorted Z-values of PAX1_PAX9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_84824920 | 2.65 |
ENST00000523678.1
|
RP11-120I21.2
|
RP11-120I21.2 |
| chr4_+_123747834 | 2.02 |
ENST00000264498.3
|
FGF2
|
fibroblast growth factor 2 (basic) |
| chr8_-_125384927 | 1.89 |
ENST00000297632.6
|
TMEM65
|
transmembrane protein 65 |
| chr4_+_123747979 | 1.77 |
ENST00000608478.1
|
FGF2
|
fibroblast growth factor 2 (basic) |
| chr6_+_34857019 | 1.46 |
ENST00000360359.3
ENST00000535627.1 |
ANKS1A
|
ankyrin repeat and sterile alpha motif domain containing 1A |
| chr9_-_124991124 | 1.43 |
ENST00000394319.4
ENST00000340587.3 |
LHX6
|
LIM homeobox 6 |
| chr19_-_18995029 | 1.22 |
ENST00000596048.1
|
CERS1
|
ceramide synthase 1 |
| chr5_-_133747589 | 1.07 |
ENST00000458198.2
|
CDKN2AIPNL
|
CDKN2A interacting protein N-terminal like |
| chr13_+_21141270 | 0.72 |
ENST00000319980.6
ENST00000537103.1 ENST00000389373.3 |
IFT88
|
intraflagellar transport 88 homolog (Chlamydomonas) |
| chr13_+_21141208 | 0.65 |
ENST00000351808.5
|
IFT88
|
intraflagellar transport 88 homolog (Chlamydomonas) |
| chr17_-_39968406 | 0.43 |
ENST00000393928.1
|
LEPREL4
|
leprecan-like 4 |
| chr6_+_132891461 | 0.41 |
ENST00000275198.1
|
TAAR6
|
trace amine associated receptor 6 |
| chr17_-_39968855 | 0.39 |
ENST00000355468.3
ENST00000590496.1 |
LEPREL4
|
leprecan-like 4 |
| chr15_+_63569785 | 0.38 |
ENST00000380343.4
ENST00000560353.1 |
APH1B
|
APH1B gamma secretase subunit |
| chr17_+_76374714 | 0.38 |
ENST00000262764.6
ENST00000589689.1 ENST00000329897.7 ENST00000592043.1 ENST00000587356.1 |
PGS1
|
phosphatidylglycerophosphate synthase 1 |
| chr15_+_63569731 | 0.31 |
ENST00000261879.5
|
APH1B
|
APH1B gamma secretase subunit |
| chr4_-_163085141 | 0.28 |
ENST00000427802.2
ENST00000306100.5 |
FSTL5
|
follistatin-like 5 |
| chr2_+_119699864 | 0.26 |
ENST00000541757.1
ENST00000412481.1 |
MARCO
|
macrophage receptor with collagenous structure |
| chrX_-_153707545 | 0.24 |
ENST00000357360.4
|
LAGE3
|
L antigen family, member 3 |
| chr1_-_204165610 | 0.23 |
ENST00000367194.4
|
KISS1
|
KiSS-1 metastasis-suppressor |
| chr18_-_47017956 | 0.21 |
ENST00000584895.1
ENST00000332968.6 ENST00000580210.1 ENST00000579408.1 |
RPL17-C18orf32
RPL17
|
RPL17-C18orf32 readthrough ribosomal protein L17 |
| chr2_-_207629997 | 0.21 |
ENST00000454776.2
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
| chr1_-_162838551 | 0.21 |
ENST00000367910.1
ENST00000367912.2 ENST00000367911.2 |
C1orf110
|
chromosome 1 open reading frame 110 |
| chr3_+_15247686 | 0.20 |
ENST00000253693.2
|
CAPN7
|
calpain 7 |
| chr11_+_6947720 | 0.19 |
ENST00000414517.2
|
ZNF215
|
zinc finger protein 215 |
| chr17_+_15848231 | 0.19 |
ENST00000304222.2
|
ADORA2B
|
adenosine A2b receptor |
| chr7_-_112758665 | 0.18 |
ENST00000397764.3
|
LINC00998
|
long intergenic non-protein coding RNA 998 |
| chr7_-_112758589 | 0.18 |
ENST00000413744.1
ENST00000439551.1 ENST00000441359.1 |
LINC00998
|
long intergenic non-protein coding RNA 998 |
| chr9_-_104249400 | 0.17 |
ENST00000374848.3
|
TMEM246
|
transmembrane protein 246 |
| chr6_-_137494775 | 0.17 |
ENST00000349184.4
ENST00000296980.2 ENST00000339602.3 |
IL22RA2
|
interleukin 22 receptor, alpha 2 |
| chrX_-_148669116 | 0.16 |
ENST00000243314.5
|
MAGEA9B
|
melanoma antigen family A, 9B |
| chrX_-_153707246 | 0.15 |
ENST00000407062.1
|
LAGE3
|
L antigen family, member 3 |
| chr17_-_3461092 | 0.15 |
ENST00000301365.4
ENST00000572519.1 |
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3 |
| chr19_+_14184370 | 0.15 |
ENST00000590772.1
|
hsa-mir-1199
|
hsa-mir-1199 |
| chr1_-_213031418 | 0.15 |
ENST00000356684.3
ENST00000426161.1 ENST00000424044.1 |
FLVCR1-AS1
|
FLVCR1 antisense RNA 1 (head to head) |
| chr2_-_162931052 | 0.14 |
ENST00000360534.3
|
DPP4
|
dipeptidyl-peptidase 4 |
| chr3_-_57113314 | 0.13 |
ENST00000338458.4
ENST00000468727.1 |
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr3_+_195447738 | 0.12 |
ENST00000447234.2
ENST00000320736.6 ENST00000436408.1 |
MUC20
|
mucin 20, cell surface associated |
| chr4_+_619386 | 0.12 |
ENST00000496514.1
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr11_+_6947647 | 0.11 |
ENST00000278319.5
|
ZNF215
|
zinc finger protein 215 |
| chr17_+_39969183 | 0.10 |
ENST00000321562.4
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr3_-_25824872 | 0.08 |
ENST00000308710.5
|
NGLY1
|
N-glycanase 1 |
| chr9_-_21142144 | 0.07 |
ENST00000380229.2
|
IFNW1
|
interferon, omega 1 |
| chr3_+_31574189 | 0.07 |
ENST00000295770.2
|
STT3B
|
STT3B, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr1_+_160121356 | 0.07 |
ENST00000368081.4
|
ATP1A4
|
ATPase, Na+/K+ transporting, alpha 4 polypeptide |
| chr11_+_61522844 | 0.06 |
ENST00000265460.5
|
MYRF
|
myelin regulatory factor |
| chr3_-_25824925 | 0.06 |
ENST00000396649.3
ENST00000428257.1 ENST00000280700.5 |
NGLY1
|
N-glycanase 1 |
| chr4_+_619347 | 0.06 |
ENST00000255622.6
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr22_+_39745930 | 0.05 |
ENST00000318801.4
ENST00000216155.7 ENST00000406293.3 ENST00000328933.5 |
SYNGR1
|
synaptogyrin 1 |
| chr4_-_77997126 | 0.05 |
ENST00000537948.1
ENST00000507788.1 ENST00000237654.4 |
CCNI
|
cyclin I |
| chr10_-_99393242 | 0.05 |
ENST00000370635.3
ENST00000478953.1 ENST00000335628.3 |
MORN4
|
MORN repeat containing 4 |
| chr6_-_170893742 | 0.05 |
ENST00000443345.2
ENST00000541970.1 |
PDCD2
|
programmed cell death 2 |
| chr14_+_24540046 | 0.04 |
ENST00000397016.2
ENST00000537691.1 ENST00000560356.1 ENST00000558450.1 |
CPNE6
|
copine VI (neuronal) |
| chr17_-_7137582 | 0.04 |
ENST00000575756.1
ENST00000575458.1 |
DVL2
|
dishevelled segment polarity protein 2 |
| chr19_-_52148798 | 0.04 |
ENST00000534261.2
|
SIGLEC5
|
sialic acid binding Ig-like lectin 5 |
| chr19_-_5838768 | 0.03 |
ENST00000527106.1
ENST00000531199.1 ENST00000529165.1 |
FUT6
|
fucosyltransferase 6 (alpha (1,3) fucosyltransferase) |
| chr8_-_121824374 | 0.02 |
ENST00000517992.1
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr7_+_142457315 | 0.02 |
ENST00000486171.1
ENST00000311737.7 |
PRSS1
|
protease, serine, 1 (trypsin 1) |
| chr22_-_41940404 | 0.01 |
ENST00000355209.4
ENST00000337566.5 ENST00000396504.2 ENST00000407461.1 |
POLR3H
|
polymerase (RNA) III (DNA directed) polypeptide H (22.9kD) |
| chr9_-_131418944 | 0.01 |
ENST00000419989.1
ENST00000451652.1 ENST00000372715.2 |
WDR34
|
WD repeat domain 34 |
| chr6_+_3118926 | 0.00 |
ENST00000380379.5
|
BPHL
|
biphenyl hydrolase-like (serine hydrolase) |
| chr18_-_47018769 | 0.00 |
ENST00000583637.1
ENST00000578528.1 ENST00000578532.1 ENST00000580387.1 ENST00000579248.1 ENST00000581373.1 |
RPL17
|
ribosomal protein L17 |
| chr17_+_37356555 | 0.00 |
ENST00000579374.1
|
RPL19
|
ribosomal protein L19 |
| chr7_-_144533074 | 0.00 |
ENST00000360057.3
ENST00000378099.3 |
TPK1
|
thiamin pyrophosphokinase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX1_PAX9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.8 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.4 | 1.2 | GO:0072720 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.4 | 1.5 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 1.4 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.2 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.2 | GO:0002880 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.0 | 0.4 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.0 | 0.7 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.1 | GO:0010716 | negative regulation of extracellular matrix disassembly(GO:0010716) |
| 0.0 | 1.4 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.2 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.0 | 2.0 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.4 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 1.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.9 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 1.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.2 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.4 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.8 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.7 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |