Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
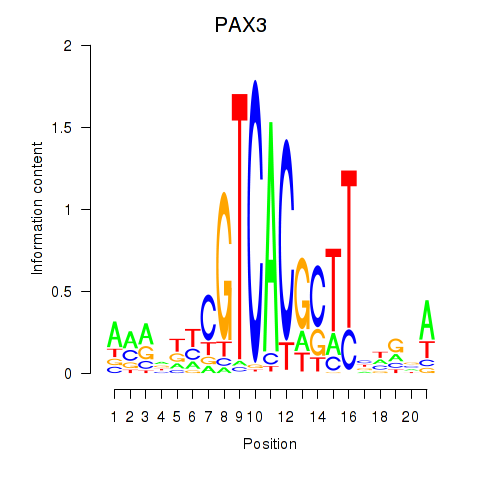
Results for PAX3
Z-value: 0.66
Transcription factors associated with PAX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX3
|
ENSG00000135903.14 | paired box 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX3 | hg19_v2_chr2_-_223163465_223163730 | 0.03 | 8.7e-01 | Click! |
Activity profile of PAX3 motif
Sorted Z-values of PAX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_101185290 | 3.91 |
ENST00000370119.4
ENST00000347652.2 ENST00000294728.2 ENST00000370115.1 |
VCAM1
|
vascular cell adhesion molecule 1 |
| chr2_+_152214098 | 3.50 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr10_+_91087651 | 2.27 |
ENST00000371818.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr15_-_80263506 | 2.07 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr6_+_14117872 | 1.51 |
ENST00000379153.3
|
CD83
|
CD83 molecule |
| chr10_-_24770632 | 1.15 |
ENST00000596413.1
|
AL353583.1
|
AL353583.1 |
| chr12_-_89919965 | 1.04 |
ENST00000548729.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr11_-_615570 | 1.03 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr4_+_8582287 | 0.99 |
ENST00000382487.4
|
GPR78
|
G protein-coupled receptor 78 |
| chr5_-_35938674 | 0.95 |
ENST00000397366.1
ENST00000513623.1 ENST00000514524.1 ENST00000397367.2 |
CAPSL
|
calcyphosine-like |
| chr2_-_11810284 | 0.94 |
ENST00000306928.5
|
NTSR2
|
neurotensin receptor 2 |
| chr11_+_313503 | 0.90 |
ENST00000528780.1
ENST00000328221.5 |
IFITM1
|
interferon induced transmembrane protein 1 |
| chr12_-_89920030 | 0.90 |
ENST00000413530.1
ENST00000547474.1 |
GALNT4
POC1B-GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) POC1B-GALNT4 readthrough |
| chr18_+_23806437 | 0.85 |
ENST00000578121.1
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa |
| chr12_+_112563335 | 0.82 |
ENST00000549358.1
ENST00000257604.5 ENST00000548092.1 ENST00000552896.1 |
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr12_+_112563303 | 0.81 |
ENST00000412615.2
|
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chrX_+_30265256 | 0.80 |
ENST00000397548.2
|
MAGEB1
|
melanoma antigen family B, 1 |
| chr22_-_44258360 | 0.79 |
ENST00000330884.4
ENST00000249130.5 |
SULT4A1
|
sulfotransferase family 4A, member 1 |
| chrX_+_38420783 | 0.78 |
ENST00000422612.2
ENST00000286824.6 ENST00000545599.1 |
TSPAN7
|
tetraspanin 7 |
| chr15_-_43622736 | 0.78 |
ENST00000544735.1
ENST00000567039.1 ENST00000305641.5 |
LCMT2
|
leucine carboxyl methyltransferase 2 |
| chr11_-_615942 | 0.77 |
ENST00000397562.3
ENST00000330243.5 ENST00000397570.1 ENST00000397574.2 |
IRF7
|
interferon regulatory factor 7 |
| chr19_+_55851221 | 0.76 |
ENST00000255613.3
ENST00000539076.1 |
SUV420H2
AC020922.1
|
suppressor of variegation 4-20 homolog 2 (Drosophila) Uncharacterized protein |
| chr4_-_170924888 | 0.72 |
ENST00000502832.1
ENST00000393704.3 |
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr3_+_14716606 | 0.70 |
ENST00000253697.3
ENST00000435614.1 ENST00000412910.1 |
C3orf20
|
chromosome 3 open reading frame 20 |
| chr21_+_34775181 | 0.66 |
ENST00000290219.6
|
IFNGR2
|
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr5_+_49961727 | 0.65 |
ENST00000505697.2
ENST00000503750.2 ENST00000514342.2 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr7_-_103629963 | 0.65 |
ENST00000428762.1
ENST00000343529.5 ENST00000424685.2 |
RELN
|
reelin |
| chr17_-_39280419 | 0.60 |
ENST00000394014.1
|
KRTAP4-12
|
keratin associated protein 4-12 |
| chr1_-_153931052 | 0.60 |
ENST00000368630.3
ENST00000368633.1 |
CRTC2
|
CREB regulated transcription coactivator 2 |
| chr19_+_38085731 | 0.59 |
ENST00000589117.1
|
ZNF540
|
zinc finger protein 540 |
| chr8_+_104426942 | 0.54 |
ENST00000297579.5
|
DCAF13
|
DDB1 and CUL4 associated factor 13 |
| chr19_-_41388657 | 0.53 |
ENST00000301146.4
ENST00000291764.3 |
CYP2A7
|
cytochrome P450, family 2, subfamily A, polypeptide 7 |
| chr19_-_48048518 | 0.53 |
ENST00000595558.1
ENST00000263351.5 |
ZNF541
|
zinc finger protein 541 |
| chr17_+_3118915 | 0.53 |
ENST00000304094.1
|
OR1A1
|
olfactory receptor, family 1, subfamily A, member 1 |
| chr2_-_114514181 | 0.51 |
ENST00000409342.1
|
SLC35F5
|
solute carrier family 35, member F5 |
| chr10_+_118305435 | 0.51 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase |
| chr1_-_205180664 | 0.46 |
ENST00000367161.3
ENST00000367162.3 ENST00000367160.4 |
DSTYK
|
dual serine/threonine and tyrosine protein kinase |
| chr6_-_169654139 | 0.46 |
ENST00000366787.3
|
THBS2
|
thrombospondin 2 |
| chr19_+_52693259 | 0.46 |
ENST00000322088.6
ENST00000454220.2 ENST00000444322.2 ENST00000477989.1 |
PPP2R1A
|
protein phosphatase 2, regulatory subunit A, alpha |
| chr6_+_89674246 | 0.45 |
ENST00000369474.1
|
AL079342.1
|
Uncharacterized protein; cDNA FLJ27030 fis, clone SLV07741 |
| chr21_-_37451680 | 0.44 |
ENST00000399201.1
|
SETD4
|
SET domain containing 4 |
| chr13_+_113623509 | 0.43 |
ENST00000535094.2
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr3_-_58572760 | 0.43 |
ENST00000447756.2
|
FAM107A
|
family with sequence similarity 107, member A |
| chr9_-_73029540 | 0.42 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr8_+_117950422 | 0.40 |
ENST00000378279.3
|
AARD
|
alanine and arginine rich domain containing protein |
| chr14_-_64804814 | 0.39 |
ENST00000554572.1
ENST00000358599.5 |
ESR2
|
estrogen receptor 2 (ER beta) |
| chr17_-_60883993 | 0.38 |
ENST00000583803.1
ENST00000456609.2 |
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr1_-_11036272 | 0.38 |
ENST00000520253.1
|
C1orf127
|
chromosome 1 open reading frame 127 |
| chr1_-_36851489 | 0.38 |
ENST00000373130.3
ENST00000373132.3 |
STK40
|
serine/threonine kinase 40 |
| chr2_-_25194476 | 0.37 |
ENST00000534855.1
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr2_+_183982238 | 0.37 |
ENST00000442895.2
ENST00000446612.1 ENST00000409798.1 |
NUP35
|
nucleoporin 35kDa |
| chr1_+_171283331 | 0.36 |
ENST00000367749.3
|
FMO4
|
flavin containing monooxygenase 4 |
| chr11_-_104972158 | 0.36 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr22_+_39101728 | 0.35 |
ENST00000216044.5
ENST00000484657.1 |
GTPBP1
|
GTP binding protein 1 |
| chrX_+_37850026 | 0.35 |
ENST00000341016.3
|
CXorf27
|
chromosome X open reading frame 27 |
| chr6_+_31939608 | 0.35 |
ENST00000375331.2
ENST00000375333.2 |
STK19
|
serine/threonine kinase 19 |
| chr1_-_36851475 | 0.35 |
ENST00000373129.3
|
STK40
|
serine/threonine kinase 40 |
| chr8_+_145159376 | 0.34 |
ENST00000322428.5
|
MAF1
|
MAF1 homolog (S. cerevisiae) |
| chrX_+_38420623 | 0.34 |
ENST00000378482.2
|
TSPAN7
|
tetraspanin 7 |
| chr8_+_145159415 | 0.34 |
ENST00000534585.1
|
MAF1
|
MAF1 homolog (S. cerevisiae) |
| chr15_-_83621435 | 0.32 |
ENST00000450735.2
ENST00000426485.1 ENST00000399166.2 ENST00000304231.8 |
HOMER2
|
homer homolog 2 (Drosophila) |
| chr11_+_31391381 | 0.32 |
ENST00000465995.1
ENST00000536040.1 |
DNAJC24
|
DnaJ (Hsp40) homolog, subfamily C, member 24 |
| chr9_+_33795533 | 0.31 |
ENST00000379405.3
|
PRSS3
|
protease, serine, 3 |
| chr1_-_205091115 | 0.30 |
ENST00000264515.6
ENST00000367164.1 |
RBBP5
|
retinoblastoma binding protein 5 |
| chr20_-_44539538 | 0.30 |
ENST00000372420.1
|
PLTP
|
phospholipid transfer protein |
| chr2_-_25194963 | 0.29 |
ENST00000264711.2
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr10_-_106098162 | 0.29 |
ENST00000337478.1
|
ITPRIP
|
inositol 1,4,5-trisphosphate receptor interacting protein |
| chr4_+_128982490 | 0.29 |
ENST00000394288.3
ENST00000432347.2 ENST00000264584.5 ENST00000441387.1 ENST00000427266.1 ENST00000354456.3 |
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr1_+_203444887 | 0.29 |
ENST00000343110.2
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein |
| chr4_+_156680153 | 0.29 |
ENST00000502959.1
ENST00000505764.1 ENST00000507146.1 ENST00000264424.8 ENST00000503520.1 |
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr2_+_183989157 | 0.28 |
ENST00000541912.1
|
NUP35
|
nucleoporin 35kDa |
| chr19_+_35168547 | 0.28 |
ENST00000502743.1
ENST00000509528.1 ENST00000506901.1 |
ZNF302
|
zinc finger protein 302 |
| chr4_+_14113592 | 0.27 |
ENST00000502759.1
ENST00000511200.1 ENST00000512754.1 ENST00000506739.1 |
LINC01085
|
long intergenic non-protein coding RNA 1085 |
| chrX_+_153770421 | 0.26 |
ENST00000369609.5
ENST00000369607.1 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr15_-_42783303 | 0.26 |
ENST00000565380.1
ENST00000564754.1 |
ZNF106
|
zinc finger protein 106 |
| chr3_-_110612323 | 0.26 |
ENST00000383686.2
|
RP11-553A10.1
|
Uncharacterized protein |
| chr9_+_131549483 | 0.24 |
ENST00000372648.5
ENST00000539497.1 |
TBC1D13
|
TBC1 domain family, member 13 |
| chr5_-_87516448 | 0.24 |
ENST00000511218.1
|
TMEM161B
|
transmembrane protein 161B |
| chr9_-_114521783 | 0.24 |
ENST00000394779.3
ENST00000394777.4 |
C9orf84
|
chromosome 9 open reading frame 84 |
| chr4_-_168155577 | 0.24 |
ENST00000541354.1
ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr15_+_45722727 | 0.24 |
ENST00000396650.2
ENST00000558435.1 ENST00000344300.3 |
C15orf48
|
chromosome 15 open reading frame 48 |
| chr6_-_9939552 | 0.24 |
ENST00000460363.2
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chrX_+_47077632 | 0.23 |
ENST00000457458.2
|
CDK16
|
cyclin-dependent kinase 16 |
| chr10_+_96162242 | 0.23 |
ENST00000225235.4
|
TBC1D12
|
TBC1 domain family, member 12 |
| chr17_+_4855053 | 0.23 |
ENST00000518175.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr4_-_168155730 | 0.22 |
ENST00000502330.1
ENST00000357154.3 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr6_-_32908765 | 0.22 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr1_+_158323244 | 0.22 |
ENST00000434258.1
|
CD1E
|
CD1e molecule |
| chr7_+_142031986 | 0.22 |
ENST00000547918.2
|
TRBV7-1
|
T cell receptor beta variable 7-1 (non-functional) |
| chr4_-_168155417 | 0.22 |
ENST00000511269.1
ENST00000506697.1 ENST00000512042.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr7_+_29519486 | 0.22 |
ENST00000409041.4
|
CHN2
|
chimerin 2 |
| chr14_+_20937538 | 0.21 |
ENST00000361505.5
ENST00000553591.1 |
PNP
|
purine nucleoside phosphorylase |
| chr2_+_219524379 | 0.21 |
ENST00000443791.1
ENST00000359273.3 ENST00000392109.1 ENST00000392110.2 ENST00000423377.1 ENST00000392111.2 ENST00000412366.1 |
BCS1L
|
BC1 (ubiquinol-cytochrome c reductase) synthesis-like |
| chr1_+_26496362 | 0.21 |
ENST00000374266.5
ENST00000270812.5 |
ZNF593
|
zinc finger protein 593 |
| chr1_+_158323486 | 0.21 |
ENST00000444681.2
ENST00000368167.3 |
CD1E
|
CD1e molecule |
| chr2_+_183989083 | 0.20 |
ENST00000295119.4
|
NUP35
|
nucleoporin 35kDa |
| chr2_-_219524193 | 0.20 |
ENST00000450560.1
ENST00000449707.1 ENST00000432460.1 ENST00000411696.2 |
ZNF142
|
zinc finger protein 142 |
| chr1_+_50513686 | 0.19 |
ENST00000448907.2
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chrX_-_73834449 | 0.19 |
ENST00000332687.6
ENST00000349225.2 |
RLIM
|
ring finger protein, LIM domain interacting |
| chr22_+_20105259 | 0.18 |
ENST00000416427.1
ENST00000421656.1 ENST00000423859.1 ENST00000418705.2 |
RANBP1
|
RAN binding protein 1 |
| chr14_-_24711806 | 0.18 |
ENST00000540705.1
ENST00000538777.1 ENST00000558566.1 ENST00000559019.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr7_-_30066233 | 0.18 |
ENST00000222803.5
|
FKBP14
|
FK506 binding protein 14, 22 kDa |
| chr12_+_10460549 | 0.18 |
ENST00000543420.1
ENST00000543777.1 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr9_+_104161123 | 0.18 |
ENST00000374861.3
ENST00000339664.2 ENST00000259395.4 |
ZNF189
|
zinc finger protein 189 |
| chrX_+_68725084 | 0.18 |
ENST00000252338.4
|
FAM155B
|
family with sequence similarity 155, member B |
| chr18_-_59274139 | 0.17 |
ENST00000586949.1
|
RP11-879F14.2
|
RP11-879F14.2 |
| chr12_-_69080590 | 0.17 |
ENST00000433116.2
ENST00000500695.2 |
RP11-637A17.2
|
RP11-637A17.2 |
| chr1_-_7913089 | 0.17 |
ENST00000361696.5
|
UTS2
|
urotensin 2 |
| chr13_+_96085847 | 0.17 |
ENST00000376873.3
|
CLDN10
|
claudin 10 |
| chr3_-_119379427 | 0.16 |
ENST00000264231.3
ENST00000468801.1 ENST00000538678.1 |
POPDC2
|
popeye domain containing 2 |
| chr11_+_73358594 | 0.16 |
ENST00000227214.6
ENST00000398494.4 ENST00000543085.1 |
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr9_+_131549610 | 0.16 |
ENST00000223865.8
|
TBC1D13
|
TBC1 domain family, member 13 |
| chr11_+_9482551 | 0.16 |
ENST00000438144.2
ENST00000526657.1 ENST00000299606.2 ENST00000534265.1 ENST00000412390.2 |
ZNF143
|
zinc finger protein 143 |
| chr1_+_99729813 | 0.16 |
ENST00000457765.1
|
LPPR4
|
Lipid phosphate phosphatase-related protein type 4 |
| chr1_-_168464875 | 0.16 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr19_-_35719609 | 0.16 |
ENST00000324675.3
|
FAM187B
|
family with sequence similarity 187, member B |
| chr19_+_7049332 | 0.15 |
ENST00000381393.3
|
MBD3L2
|
methyl-CpG binding domain protein 3-like 2 |
| chr8_-_10512569 | 0.15 |
ENST00000382483.3
|
RP1L1
|
retinitis pigmentosa 1-like 1 |
| chr9_-_91793675 | 0.15 |
ENST00000375835.4
ENST00000375830.1 |
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr10_+_134150835 | 0.15 |
ENST00000432555.2
|
LRRC27
|
leucine rich repeat containing 27 |
| chr19_-_2783363 | 0.14 |
ENST00000221566.2
|
SGTA
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr16_+_27214802 | 0.14 |
ENST00000380948.2
ENST00000286096.4 |
KDM8
|
lysine (K)-specific demethylase 8 |
| chr7_+_99816859 | 0.14 |
ENST00000317271.2
|
PVRIG
|
poliovirus receptor related immunoglobulin domain containing |
| chr17_-_39538550 | 0.14 |
ENST00000394001.1
|
KRT34
|
keratin 34 |
| chr4_-_168155700 | 0.14 |
ENST00000357545.4
ENST00000512648.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr16_+_23847339 | 0.13 |
ENST00000303531.7
|
PRKCB
|
protein kinase C, beta |
| chr15_+_51973550 | 0.13 |
ENST00000220478.3
|
SCG3
|
secretogranin III |
| chr1_-_145715565 | 0.13 |
ENST00000369288.2
ENST00000369290.1 ENST00000401557.3 |
CD160
|
CD160 molecule |
| chrX_+_48398053 | 0.13 |
ENST00000537536.1
ENST00000418627.1 |
TBC1D25
|
TBC1 domain family, member 25 |
| chr5_-_179498703 | 0.13 |
ENST00000522208.2
|
RNF130
|
ring finger protein 130 |
| chr3_+_38307293 | 0.12 |
ENST00000311856.4
|
SLC22A13
|
solute carrier family 22 (organic anion/urate transporter), member 13 |
| chr4_+_156680143 | 0.12 |
ENST00000505154.1
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr2_-_68479614 | 0.12 |
ENST00000234310.3
|
PPP3R1
|
protein phosphatase 3, regulatory subunit B, alpha |
| chrX_-_70326455 | 0.12 |
ENST00000374251.5
|
CXorf65
|
chromosome X open reading frame 65 |
| chr1_+_40505891 | 0.11 |
ENST00000372797.3
ENST00000372802.1 ENST00000449311.1 |
CAP1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr5_+_178977546 | 0.11 |
ENST00000319449.4
ENST00000377001.2 |
RUFY1
|
RUN and FYVE domain containing 1 |
| chr3_+_49236910 | 0.11 |
ENST00000452691.2
ENST00000366429.2 |
CCDC36
|
coiled-coil domain containing 36 |
| chr15_+_51973680 | 0.11 |
ENST00000542355.2
|
SCG3
|
secretogranin III |
| chr19_+_38085768 | 0.11 |
ENST00000316433.4
ENST00000590588.1 ENST00000586134.1 ENST00000586792.1 |
ZNF540
|
zinc finger protein 540 |
| chr1_+_202830876 | 0.11 |
ENST00000456105.2
|
RP11-480I12.7
|
RP11-480I12.7 |
| chr21_-_30365136 | 0.10 |
ENST00000361371.5
ENST00000389194.2 ENST00000389195.2 |
LTN1
|
listerin E3 ubiquitin protein ligase 1 |
| chr6_-_109804412 | 0.10 |
ENST00000230122.3
|
ZBTB24
|
zinc finger and BTB domain containing 24 |
| chr10_-_102289611 | 0.10 |
ENST00000299166.4
ENST00000370320.4 ENST00000531258.1 ENST00000370322.1 ENST00000535773.1 |
NDUFB8
SEC31B
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8, 19kDa SEC31 homolog B (S. cerevisiae) |
| chr3_+_127770455 | 0.09 |
ENST00000464451.1
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr19_-_46296011 | 0.09 |
ENST00000377735.3
ENST00000270223.6 |
DMWD
|
dystrophia myotonica, WD repeat containing |
| chr12_-_10562356 | 0.09 |
ENST00000309384.1
|
KLRC4
|
killer cell lectin-like receptor subfamily C, member 4 |
| chr2_+_242811874 | 0.09 |
ENST00000343216.3
|
CXXC11
|
CXXC finger protein 11 |
| chr16_-_71523179 | 0.08 |
ENST00000564230.1
ENST00000565637.1 |
ZNF19
|
zinc finger protein 19 |
| chr2_-_227050079 | 0.08 |
ENST00000423838.1
|
AC068138.1
|
AC068138.1 |
| chr12_+_10460417 | 0.08 |
ENST00000381908.3
ENST00000336164.4 ENST00000350274.5 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr15_+_81591757 | 0.08 |
ENST00000558332.1
|
IL16
|
interleukin 16 |
| chr2_-_44065889 | 0.08 |
ENST00000543989.1
ENST00000405322.1 |
ABCG5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr11_+_65122216 | 0.07 |
ENST00000309880.5
|
TIGD3
|
tigger transposable element derived 3 |
| chr19_+_2841433 | 0.07 |
ENST00000334241.4
ENST00000585966.1 ENST00000591539.1 |
ZNF555
|
zinc finger protein 555 |
| chr6_-_30815936 | 0.07 |
ENST00000442852.1
|
XXbac-BPG27H4.8
|
XXbac-BPG27H4.8 |
| chr20_-_23807358 | 0.06 |
ENST00000304725.2
|
CST2
|
cystatin SA |
| chr11_+_9482512 | 0.05 |
ENST00000396602.2
ENST00000530463.1 ENST00000533542.1 ENST00000532577.1 ENST00000396597.3 |
ZNF143
|
zinc finger protein 143 |
| chr19_+_32836499 | 0.05 |
ENST00000311921.4
ENST00000544431.1 ENST00000355898.5 |
ZNF507
|
zinc finger protein 507 |
| chrX_-_48931648 | 0.05 |
ENST00000376386.3
ENST00000376390.4 |
PRAF2
|
PRA1 domain family, member 2 |
| chr12_-_75603643 | 0.05 |
ENST00000549446.1
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr16_-_71523236 | 0.05 |
ENST00000288177.5
ENST00000569072.1 |
ZNF19
|
zinc finger protein 19 |
| chr16_-_52640834 | 0.04 |
ENST00000510238.3
|
CASC16
|
cancer susceptibility candidate 16 (non-protein coding) |
| chr8_-_145159083 | 0.04 |
ENST00000398712.2
|
SHARPIN
|
SHANK-associated RH domain interactor |
| chr19_+_35168633 | 0.04 |
ENST00000505365.2
|
ZNF302
|
zinc finger protein 302 |
| chr12_-_53594227 | 0.04 |
ENST00000550743.2
|
ITGB7
|
integrin, beta 7 |
| chr4_+_128982430 | 0.03 |
ENST00000512292.1
ENST00000508819.1 |
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr6_-_32908792 | 0.03 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr1_+_247670415 | 0.03 |
ENST00000366491.2
ENST00000366489.1 ENST00000526896.1 |
GCSAML
|
germinal center-associated, signaling and motility-like |
| chrX_+_70364667 | 0.03 |
ENST00000536169.1
ENST00000395855.2 ENST00000374051.3 ENST00000358741.3 |
NLGN3
|
neuroligin 3 |
| chr16_-_75018968 | 0.03 |
ENST00000262144.6
|
WDR59
|
WD repeat domain 59 |
| chr19_-_42463418 | 0.03 |
ENST00000600292.1
ENST00000601078.1 ENST00000601891.1 ENST00000222008.6 |
RABAC1
|
Rab acceptor 1 (prenylated) |
| chr19_+_4343691 | 0.03 |
ENST00000597036.1
|
MPND
|
MPN domain containing |
| chr7_+_142457315 | 0.03 |
ENST00000486171.1
ENST00000311737.7 |
PRSS1
|
protease, serine, 1 (trypsin 1) |
| chr4_+_15376165 | 0.03 |
ENST00000382383.3
ENST00000429690.1 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr14_+_22217447 | 0.02 |
ENST00000390427.3
|
TRAV5
|
T cell receptor alpha variable 5 |
| chr5_+_69345350 | 0.02 |
ENST00000380741.4
ENST00000380743.4 ENST00000511812.1 ENST00000380742.4 |
SMN2
|
survival of motor neuron 2, centromeric |
| chr19_-_54784353 | 0.02 |
ENST00000391746.1
|
LILRB2
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 2 |
| chr21_-_31744557 | 0.02 |
ENST00000399889.2
|
KRTAP13-2
|
keratin associated protein 13-2 |
| chr8_-_101718991 | 0.02 |
ENST00000517990.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr12_-_127544894 | 0.02 |
ENST00000546062.1
ENST00000512624.2 ENST00000540244.1 |
RP11-575F12.1
|
RP11-575F12.1 |
| chr8_+_125463048 | 0.02 |
ENST00000328599.3
|
TRMT12
|
tRNA methyltransferase 12 homolog (S. cerevisiae) |
| chr1_-_153123345 | 0.01 |
ENST00000368748.4
|
SPRR2G
|
small proline-rich protein 2G |
| chr2_+_171034646 | 0.01 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
| chrX_-_13835461 | 0.01 |
ENST00000316715.4
ENST00000356942.5 |
GPM6B
|
glycoprotein M6B |
| chr12_+_652294 | 0.01 |
ENST00000322843.3
|
B4GALNT3
|
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
| chr17_-_3461092 | 0.00 |
ENST00000301365.4
ENST00000572519.1 |
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3 |
| chr5_+_70220768 | 0.00 |
ENST00000351205.4
ENST00000503079.2 ENST00000380707.4 ENST00000514951.1 ENST00000506163.1 |
SMN1
|
survival of motor neuron 1, telomeric |
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.9 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.4 | 1.8 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.2 | 2.3 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.2 | 1.5 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.2 | 0.6 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.7 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.5 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.7 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 3.5 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 0.4 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.3 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.1 | 0.3 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.1 | 0.2 | GO:0046495 | nicotinamide riboside catabolic process(GO:0006738) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.1 | 2.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.5 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.4 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.1 | 0.5 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 | 0.2 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 0.9 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.4 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.8 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.6 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.0 | 0.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.2 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.3 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.5 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.3 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.7 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.6 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.0 | 0.2 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 1.6 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.7 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.4 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.8 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.8 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.2 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.3 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.8 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.4 | GO:1900116 | extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.3 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.9 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.2 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 0.4 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) protease inhibitor complex(GO:0097179) |
| 0.1 | 0.9 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.2 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 3.2 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.7 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 0.7 | GO:0000994 | RNA polymerase III core binding(GO:0000994) |
| 0.2 | 0.6 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.1 | 0.7 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.1 | 3.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 2.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.2 | GO:0000035 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.2 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.1 | 0.3 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.8 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.4 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.0 | 0.4 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.4 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.5 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.8 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.7 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.9 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.7 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 1.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 4.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.8 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 0.7 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 5.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.7 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.5 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |