Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
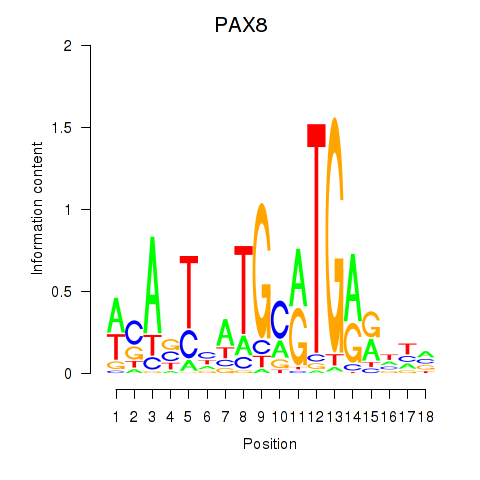
Results for PAX8
Z-value: 0.92
Transcription factors associated with PAX8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX8
|
ENSG00000125618.12 | paired box 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX8 | hg19_v2_chr2_-_113999260_113999274 | -0.42 | 3.7e-02 | Click! |
Activity profile of PAX8 motif
Sorted Z-values of PAX8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_169703203 | 7.14 |
ENST00000333360.7
ENST00000367781.4 ENST00000367782.4 ENST00000367780.4 ENST00000367779.4 |
SELE
|
selectin E |
| chr6_-_44233361 | 3.05 |
ENST00000275015.5
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| chr11_+_20044600 | 2.84 |
ENST00000311043.8
|
NAV2
|
neuron navigator 2 |
| chr15_+_67420441 | 2.71 |
ENST00000558894.1
|
SMAD3
|
SMAD family member 3 |
| chr14_-_91710852 | 2.60 |
ENST00000535815.1
ENST00000529102.1 |
GPR68
|
G protein-coupled receptor 68 |
| chr1_-_173176452 | 2.48 |
ENST00000281834.3
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chr11_+_20044096 | 2.28 |
ENST00000533917.1
|
NAV2
|
neuron navigator 2 |
| chr6_+_14117872 | 1.91 |
ENST00000379153.3
|
CD83
|
CD83 molecule |
| chr6_-_134639180 | 1.72 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr12_+_26348582 | 1.66 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr17_+_80332153 | 1.64 |
ENST00000313135.2
|
UTS2R
|
urotensin 2 receptor |
| chr16_+_66442411 | 1.55 |
ENST00000499966.1
|
LINC00920
|
long intergenic non-protein coding RNA 920 |
| chr22_+_21133469 | 1.54 |
ENST00000406799.1
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr6_-_82462425 | 1.44 |
ENST00000369754.3
ENST00000320172.6 ENST00000369756.3 |
FAM46A
|
family with sequence similarity 46, member A |
| chr12_-_129308487 | 1.37 |
ENST00000266771.5
|
SLC15A4
|
solute carrier family 15 (oligopeptide transporter), member 4 |
| chr12_-_89919965 | 1.36 |
ENST00000548729.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr3_+_11178779 | 1.30 |
ENST00000438284.2
|
HRH1
|
histamine receptor H1 |
| chr14_+_22458631 | 1.23 |
ENST00000390444.1
|
TRAV16
|
T cell receptor alpha variable 16 |
| chr1_-_231004220 | 1.20 |
ENST00000366663.5
|
C1orf198
|
chromosome 1 open reading frame 198 |
| chr6_+_29274403 | 1.19 |
ENST00000377160.2
|
OR14J1
|
olfactory receptor, family 14, subfamily J, member 1 |
| chr22_+_21128167 | 1.17 |
ENST00000215727.5
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr12_-_56236734 | 1.11 |
ENST00000548629.1
|
MMP19
|
matrix metallopeptidase 19 |
| chr4_-_69536346 | 1.06 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr3_-_189840223 | 1.05 |
ENST00000427335.2
|
LEPREL1
|
leprecan-like 1 |
| chr10_+_85933494 | 0.98 |
ENST00000372126.3
|
C10orf99
|
chromosome 10 open reading frame 99 |
| chr12_+_75874460 | 0.98 |
ENST00000266659.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr7_+_128312346 | 0.92 |
ENST00000480462.1
ENST00000378704.3 ENST00000477515.1 |
FAM71F2
|
family with sequence similarity 71, member F2 |
| chr2_+_161993465 | 0.90 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr12_+_72058130 | 0.90 |
ENST00000547843.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr9_+_111624577 | 0.89 |
ENST00000333999.3
|
ACTL7A
|
actin-like 7A |
| chr1_+_159409512 | 0.89 |
ENST00000423932.3
|
OR10J1
|
olfactory receptor, family 10, subfamily J, member 1 |
| chr1_+_87012753 | 0.86 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chr10_+_70847852 | 0.85 |
ENST00000242465.3
|
SRGN
|
serglycin |
| chr1_+_145293371 | 0.85 |
ENST00000342960.5
|
NBPF10
|
neuroblastoma breakpoint family, member 10 |
| chr10_+_103912137 | 0.83 |
ENST00000603742.1
ENST00000488254.2 ENST00000461421.1 ENST00000476468.1 ENST00000370007.5 |
NOLC1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr15_+_89631647 | 0.82 |
ENST00000569550.1
ENST00000565066.1 ENST00000565973.1 |
ABHD2
|
abhydrolase domain containing 2 |
| chr11_-_104905840 | 0.81 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr4_+_36283213 | 0.80 |
ENST00000357504.3
|
DTHD1
|
death domain containing 1 |
| chr7_-_97501706 | 0.77 |
ENST00000455086.1
ENST00000453600.1 |
ASNS
|
asparagine synthetase (glutamine-hydrolyzing) |
| chr8_+_120079478 | 0.76 |
ENST00000332843.2
|
COLEC10
|
collectin sub-family member 10 (C-type lectin) |
| chr3_-_107941209 | 0.75 |
ENST00000492106.1
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr4_-_23735183 | 0.73 |
ENST00000507666.1
|
RP11-380P13.2
|
RP11-380P13.2 |
| chr11_-_62609281 | 0.71 |
ENST00000525239.1
ENST00000538098.2 |
WDR74
|
WD repeat domain 74 |
| chr7_-_142176790 | 0.71 |
ENST00000390369.2
|
TRBV7-4
|
T cell receptor beta variable 7-4 (gene/pseudogene) |
| chr12_-_56236690 | 0.71 |
ENST00000322569.4
|
MMP19
|
matrix metallopeptidase 19 |
| chr2_+_185463093 | 0.70 |
ENST00000302277.6
|
ZNF804A
|
zinc finger protein 804A |
| chr8_+_31496809 | 0.69 |
ENST00000518104.1
ENST00000519301.1 |
NRG1
|
neuregulin 1 |
| chr6_+_135502501 | 0.68 |
ENST00000527615.1
ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr4_+_120860695 | 0.68 |
ENST00000505122.2
|
RP11-700N1.1
|
RP11-700N1.1 |
| chr17_-_39216344 | 0.68 |
ENST00000391418.2
|
KRTAP2-3
|
keratin associated protein 2-3 |
| chr7_-_137686791 | 0.68 |
ENST00000452463.1
ENST00000330387.6 ENST00000456390.1 |
CREB3L2
|
cAMP responsive element binding protein 3-like 2 |
| chr9_-_117853297 | 0.66 |
ENST00000542877.1
ENST00000537320.1 ENST00000341037.4 |
TNC
|
tenascin C |
| chr10_+_103911926 | 0.65 |
ENST00000605788.1
ENST00000405356.1 |
NOLC1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr17_-_60883993 | 0.65 |
ENST00000583803.1
ENST00000456609.2 |
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr11_-_47870091 | 0.64 |
ENST00000526870.1
|
NUP160
|
nucleoporin 160kDa |
| chr2_+_190306159 | 0.64 |
ENST00000314761.4
|
WDR75
|
WD repeat domain 75 |
| chr17_-_34257731 | 0.63 |
ENST00000431884.2
ENST00000425909.3 ENST00000394528.3 ENST00000430160.2 |
RDM1
|
RAD52 motif 1 |
| chr15_-_72523924 | 0.63 |
ENST00000566809.1
ENST00000567087.1 ENST00000569050.1 ENST00000568883.1 |
PKM
|
pyruvate kinase, muscle |
| chr17_+_7461849 | 0.62 |
ENST00000338784.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr2_+_161993412 | 0.62 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr6_+_135502408 | 0.61 |
ENST00000341911.5
ENST00000442647.2 ENST00000316528.8 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr8_-_20040638 | 0.61 |
ENST00000519026.1
ENST00000276373.5 ENST00000440926.1 ENST00000437980.1 |
SLC18A1
|
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr12_-_50294033 | 0.61 |
ENST00000552669.1
|
FAIM2
|
Fas apoptotic inhibitory molecule 2 |
| chr9_-_134955246 | 0.61 |
ENST00000357028.2
ENST00000474263.1 ENST00000292035.5 |
MED27
|
mediator complex subunit 27 |
| chr20_+_4152356 | 0.61 |
ENST00000379460.2
|
SMOX
|
spermine oxidase |
| chr14_+_101123580 | 0.60 |
ENST00000556697.1
ENST00000360899.2 ENST00000553623.1 |
LINC00523
|
long intergenic non-protein coding RNA 523 |
| chr13_+_27844464 | 0.59 |
ENST00000241463.4
|
RASL11A
|
RAS-like, family 11, member A |
| chr6_+_43739697 | 0.59 |
ENST00000230480.6
|
VEGFA
|
vascular endothelial growth factor A |
| chr15_-_89010607 | 0.58 |
ENST00000312475.4
|
MRPL46
|
mitochondrial ribosomal protein L46 |
| chr2_-_3606206 | 0.57 |
ENST00000315212.3
|
RNASEH1
|
ribonuclease H1 |
| chr11_+_64323156 | 0.56 |
ENST00000377585.3
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr7_-_76255444 | 0.55 |
ENST00000454397.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr22_-_32651326 | 0.55 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr19_+_40476912 | 0.55 |
ENST00000157812.2
|
PSMC4
|
proteasome (prosome, macropain) 26S subunit, ATPase, 4 |
| chr12_-_10251539 | 0.54 |
ENST00000420265.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr7_+_107531580 | 0.54 |
ENST00000537148.1
ENST00000440410.1 ENST00000437604.2 |
DLD
|
dihydrolipoamide dehydrogenase |
| chr17_-_34257771 | 0.54 |
ENST00000394529.3
ENST00000293273.6 |
RDM1
|
RAD52 motif 1 |
| chr10_-_21435488 | 0.54 |
ENST00000534331.1
ENST00000529198.1 ENST00000377118.4 |
C10orf113
|
chromosome 10 open reading frame 113 |
| chr4_+_156680153 | 0.54 |
ENST00000502959.1
ENST00000505764.1 ENST00000507146.1 ENST00000264424.8 ENST00000503520.1 |
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr1_-_153514241 | 0.53 |
ENST00000368718.1
ENST00000359215.1 |
S100A5
|
S100 calcium binding protein A5 |
| chr6_+_135502466 | 0.52 |
ENST00000367814.4
|
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr12_-_10251603 | 0.52 |
ENST00000457018.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr17_+_7462103 | 0.52 |
ENST00000396545.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr3_-_161090660 | 0.51 |
ENST00000359175.4
|
SPTSSB
|
serine palmitoyltransferase, small subunit B |
| chr1_+_100818156 | 0.51 |
ENST00000336454.3
|
CDC14A
|
cell division cycle 14A |
| chr18_-_10748498 | 0.51 |
ENST00000579949.1
|
PIEZO2
|
piezo-type mechanosensitive ion channel component 2 |
| chr20_-_42355629 | 0.50 |
ENST00000373003.1
|
GTSF1L
|
gametocyte specific factor 1-like |
| chr12_+_57828521 | 0.50 |
ENST00000309668.2
|
INHBC
|
inhibin, beta C |
| chr1_-_11036272 | 0.50 |
ENST00000520253.1
|
C1orf127
|
chromosome 1 open reading frame 127 |
| chr3_-_150264272 | 0.49 |
ENST00000491660.1
ENST00000487153.1 ENST00000239944.2 |
SERP1
|
stress-associated endoplasmic reticulum protein 1 |
| chr2_+_79252822 | 0.49 |
ENST00000272324.5
|
REG3G
|
regenerating islet-derived 3 gamma |
| chr9_-_128246769 | 0.49 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr5_+_69321074 | 0.48 |
ENST00000380751.5
ENST00000380750.3 ENST00000503931.1 ENST00000506542.1 |
SERF1B
|
small EDRK-rich factor 1B (centromeric) |
| chr17_+_7462031 | 0.48 |
ENST00000380535.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr17_+_7461781 | 0.48 |
ENST00000349228.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr15_-_43785274 | 0.48 |
ENST00000413546.1
|
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr3_+_121796697 | 0.48 |
ENST00000482356.1
ENST00000393627.2 |
CD86
|
CD86 molecule |
| chr6_+_30130969 | 0.48 |
ENST00000376694.4
|
TRIM15
|
tripartite motif containing 15 |
| chr19_+_35630926 | 0.47 |
ENST00000588081.1
ENST00000589121.1 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr12_-_10251576 | 0.47 |
ENST00000315330.4
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr7_+_7606497 | 0.46 |
ENST00000340080.4
ENST00000405785.1 ENST00000433635.1 |
MIOS
|
missing oocyte, meiosis regulator, homolog (Drosophila) |
| chr3_-_110612323 | 0.46 |
ENST00000383686.2
|
RP11-553A10.1
|
Uncharacterized protein |
| chr6_+_131571535 | 0.46 |
ENST00000474850.2
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr2_+_90259593 | 0.46 |
ENST00000471857.1
|
IGKV1D-8
|
immunoglobulin kappa variable 1D-8 |
| chr17_+_7461613 | 0.46 |
ENST00000438470.1
ENST00000436057.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr3_-_110612059 | 0.45 |
ENST00000485473.1
|
RP11-553A10.1
|
Uncharacterized protein |
| chr15_+_89631381 | 0.45 |
ENST00000352732.5
|
ABHD2
|
abhydrolase domain containing 2 |
| chr4_+_156680143 | 0.45 |
ENST00000505154.1
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr6_-_9939552 | 0.44 |
ENST00000460363.2
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr20_+_2795626 | 0.43 |
ENST00000603872.1
ENST00000380589.4 |
C20orf141
|
chromosome 20 open reading frame 141 |
| chr2_+_79252804 | 0.43 |
ENST00000393897.2
|
REG3G
|
regenerating islet-derived 3 gamma |
| chr2_+_85661918 | 0.42 |
ENST00000340326.2
|
SH2D6
|
SH2 domain containing 6 |
| chr6_+_31126291 | 0.41 |
ENST00000376257.3
ENST00000376255.4 |
TCF19
|
transcription factor 19 |
| chr6_+_160542870 | 0.41 |
ENST00000324965.4
ENST00000457470.2 |
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr10_+_24738355 | 0.39 |
ENST00000307544.6
|
KIAA1217
|
KIAA1217 |
| chr1_+_197382957 | 0.39 |
ENST00000367397.1
|
CRB1
|
crumbs homolog 1 (Drosophila) |
| chr1_+_180165672 | 0.38 |
ENST00000443059.1
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr16_+_57496299 | 0.37 |
ENST00000219252.5
|
POLR2C
|
polymerase (RNA) II (DNA directed) polypeptide C, 33kDa |
| chr6_+_160542821 | 0.37 |
ENST00000366963.4
|
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr1_+_203734296 | 0.37 |
ENST00000442561.2
ENST00000367217.5 |
LAX1
|
lymphocyte transmembrane adaptor 1 |
| chr12_-_53074182 | 0.37 |
ENST00000252244.3
|
KRT1
|
keratin 1 |
| chr9_+_125376948 | 0.37 |
ENST00000297913.2
|
OR1Q1
|
olfactory receptor, family 1, subfamily Q, member 1 |
| chr10_+_96443378 | 0.36 |
ENST00000285979.6
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18 |
| chr4_-_168155169 | 0.35 |
ENST00000534949.1
ENST00000535728.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr22_-_44258360 | 0.35 |
ENST00000330884.4
ENST00000249130.5 |
SULT4A1
|
sulfotransferase family 4A, member 1 |
| chr17_+_7461580 | 0.34 |
ENST00000483039.1
ENST00000396542.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr19_-_51537982 | 0.34 |
ENST00000525263.1
|
KLK12
|
kallikrein-related peptidase 12 |
| chr4_-_46911223 | 0.33 |
ENST00000396533.1
|
COX7B2
|
cytochrome c oxidase subunit VIIb2 |
| chr6_-_32784687 | 0.33 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr21_-_35883541 | 0.33 |
ENST00000399284.1
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1 |
| chr3_-_8811288 | 0.32 |
ENST00000316793.3
ENST00000431493.1 |
OXTR
|
oxytocin receptor |
| chr19_+_50169081 | 0.31 |
ENST00000246784.3
|
BCL2L12
|
BCL2-like 12 (proline rich) |
| chr19_-_40228657 | 0.30 |
ENST00000221804.4
|
CLC
|
Charcot-Leyden crystal galectin |
| chr6_+_24126350 | 0.30 |
ENST00000378491.4
ENST00000378478.1 ENST00000378477.2 |
NRSN1
|
neurensin 1 |
| chr12_-_6961050 | 0.30 |
ENST00000538862.2
|
CDCA3
|
cell division cycle associated 3 |
| chr12_-_18890940 | 0.30 |
ENST00000543242.1
ENST00000539072.1 ENST00000541966.1 ENST00000266505.7 ENST00000447925.2 ENST00000435379.1 |
PLCZ1
|
phospholipase C, zeta 1 |
| chr3_-_107941230 | 0.29 |
ENST00000264538.3
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr2_-_26864228 | 0.29 |
ENST00000288861.4
|
CIB4
|
calcium and integrin binding family member 4 |
| chr19_+_18794470 | 0.29 |
ENST00000321949.8
ENST00000338797.6 |
CRTC1
|
CREB regulated transcription coactivator 1 |
| chr12_-_86650045 | 0.29 |
ENST00000604798.1
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr17_-_33760269 | 0.28 |
ENST00000452764.3
|
SLFN12
|
schlafen family member 12 |
| chrX_-_23926004 | 0.28 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr19_-_52133588 | 0.28 |
ENST00000570106.2
|
SIGLEC5
|
sialic acid binding Ig-like lectin 5 |
| chr3_+_44754126 | 0.27 |
ENST00000449836.1
ENST00000436624.2 ENST00000296091.4 ENST00000411443.1 |
ZNF502
|
zinc finger protein 502 |
| chr7_+_129984630 | 0.27 |
ENST00000355388.3
ENST00000497503.1 ENST00000463587.1 ENST00000461828.1 ENST00000494311.1 ENST00000466363.2 ENST00000485477.1 ENST00000431780.2 ENST00000474905.1 |
CPA5
|
carboxypeptidase A5 |
| chr2_-_179672142 | 0.27 |
ENST00000342992.6
ENST00000360870.5 ENST00000460472.2 ENST00000589042.1 ENST00000591111.1 ENST00000342175.6 ENST00000359218.5 |
TTN
|
titin |
| chr12_-_57873631 | 0.26 |
ENST00000393791.3
ENST00000356411.2 ENST00000552249.1 |
ARHGAP9
|
Rho GTPase activating protein 9 |
| chr1_-_21059029 | 0.25 |
ENST00000444387.2
ENST00000375031.1 ENST00000518294.1 |
SH2D5
|
SH2 domain containing 5 |
| chr15_-_49255632 | 0.25 |
ENST00000332408.4
|
SHC4
|
SHC (Src homology 2 domain containing) family, member 4 |
| chr1_+_171283331 | 0.25 |
ENST00000367749.3
|
FMO4
|
flavin containing monooxygenase 4 |
| chr2_+_90192768 | 0.25 |
ENST00000390275.2
|
IGKV1D-13
|
immunoglobulin kappa variable 1D-13 |
| chr10_+_135340859 | 0.25 |
ENST00000252945.3
ENST00000421586.1 ENST00000418356.1 |
CYP2E1
|
cytochrome P450, family 2, subfamily E, polypeptide 1 |
| chr11_+_60609537 | 0.24 |
ENST00000227520.5
|
CCDC86
|
coiled-coil domain containing 86 |
| chr16_-_20367584 | 0.24 |
ENST00000570689.1
|
UMOD
|
uromodulin |
| chr15_-_43785303 | 0.24 |
ENST00000382039.3
ENST00000450115.2 ENST00000382044.4 |
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr19_+_35939154 | 0.24 |
ENST00000599180.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr10_-_43892668 | 0.24 |
ENST00000544000.1
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr1_+_155006300 | 0.24 |
ENST00000295542.1
ENST00000392480.1 ENST00000423025.2 ENST00000368419.2 |
DCST1
|
DC-STAMP domain containing 1 |
| chr2_-_79313973 | 0.24 |
ENST00000454188.1
|
REG1B
|
regenerating islet-derived 1 beta |
| chr19_-_7812446 | 0.24 |
ENST00000394173.4
ENST00000301357.8 |
CD209
|
CD209 molecule |
| chr11_+_113185292 | 0.24 |
ENST00000429951.1
ENST00000442859.1 ENST00000531164.1 ENST00000529850.1 ENST00000314756.3 ENST00000525965.1 |
TTC12
|
tetratricopeptide repeat domain 12 |
| chrY_-_13524717 | 0.23 |
ENST00000331172.6
|
SLC9B1P1
|
solute carrier family 9, subfamily B (NHA1, cation proton antiporter 1), member 1 pseudogene 1 |
| chr8_+_39792474 | 0.23 |
ENST00000502986.2
|
IDO2
|
indoleamine 2,3-dioxygenase 2 |
| chr17_-_80017856 | 0.23 |
ENST00000577574.1
|
DUS1L
|
dihydrouridine synthase 1-like (S. cerevisiae) |
| chr2_+_219524473 | 0.22 |
ENST00000439945.1
ENST00000431802.1 |
BCS1L
|
BC1 (ubiquinol-cytochrome c reductase) synthesis-like |
| chr4_+_119809984 | 0.22 |
ENST00000307142.4
ENST00000448416.2 ENST00000429713.2 |
SYNPO2
|
synaptopodin 2 |
| chr19_-_55791431 | 0.22 |
ENST00000593263.1
ENST00000376343.3 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr6_-_31628512 | 0.22 |
ENST00000375911.1
|
C6orf47
|
chromosome 6 open reading frame 47 |
| chr5_-_133510456 | 0.22 |
ENST00000520417.1
|
SKP1
|
S-phase kinase-associated protein 1 |
| chr19_+_9361606 | 0.21 |
ENST00000456448.1
|
OR7E24
|
olfactory receptor, family 7, subfamily E, member 24 |
| chr4_+_75230853 | 0.21 |
ENST00000244869.2
|
EREG
|
epiregulin |
| chr1_-_157522260 | 0.20 |
ENST00000368191.3
ENST00000361835.3 |
FCRL5
|
Fc receptor-like 5 |
| chr15_-_35088340 | 0.20 |
ENST00000290378.4
|
ACTC1
|
actin, alpha, cardiac muscle 1 |
| chr11_+_67222818 | 0.20 |
ENST00000325656.5
|
CABP4
|
calcium binding protein 4 |
| chr10_-_104597286 | 0.20 |
ENST00000369887.3
|
CYP17A1
|
cytochrome P450, family 17, subfamily A, polypeptide 1 |
| chr2_-_37193606 | 0.20 |
ENST00000379213.2
ENST00000263918.4 |
STRN
|
striatin, calmodulin binding protein |
| chr21_-_43816052 | 0.19 |
ENST00000398405.1
|
TMPRSS3
|
transmembrane protease, serine 3 |
| chr8_-_53322303 | 0.19 |
ENST00000276480.7
|
ST18
|
suppression of tumorigenicity 18 (breast carcinoma) (zinc finger protein) |
| chr5_+_140235469 | 0.19 |
ENST00000506939.2
ENST00000307360.5 |
PCDHA10
|
protocadherin alpha 10 |
| chr11_+_55594695 | 0.19 |
ENST00000378397.1
|
OR5L2
|
olfactory receptor, family 5, subfamily L, member 2 |
| chr19_-_55791540 | 0.19 |
ENST00000433386.2
|
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr1_+_171227069 | 0.19 |
ENST00000354841.4
|
FMO1
|
flavin containing monooxygenase 1 |
| chr1_-_186649543 | 0.18 |
ENST00000367468.5
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chr4_+_71091786 | 0.18 |
ENST00000317987.5
|
FDCSP
|
follicular dendritic cell secreted protein |
| chr19_-_55791563 | 0.18 |
ENST00000588971.1
ENST00000255631.5 ENST00000587551.1 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr1_-_27952741 | 0.18 |
ENST00000399173.1
|
FGR
|
feline Gardner-Rasheed sarcoma viral oncogene homolog |
| chr19_-_51538118 | 0.18 |
ENST00000529888.1
|
KLK12
|
kallikrein-related peptidase 12 |
| chr22_+_23029188 | 0.18 |
ENST00000390305.2
|
IGLV3-25
|
immunoglobulin lambda variable 3-25 |
| chr22_-_37403858 | 0.17 |
ENST00000405091.2
|
TEX33
|
testis expressed 33 |
| chr11_-_58343319 | 0.17 |
ENST00000395074.2
|
LPXN
|
leupaxin |
| chr2_-_29297127 | 0.17 |
ENST00000331664.5
|
C2orf71
|
chromosome 2 open reading frame 71 |
| chr11_+_60223312 | 0.17 |
ENST00000532491.1
ENST00000532073.1 ENST00000534668.1 ENST00000528313.1 ENST00000533306.1 |
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr22_-_37403839 | 0.17 |
ENST00000402860.3
ENST00000381821.1 |
TEX33
|
testis expressed 33 |
| chr10_+_12391685 | 0.17 |
ENST00000378845.1
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID |
| chr17_+_6918093 | 0.17 |
ENST00000439424.2
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr1_-_157522180 | 0.16 |
ENST00000356953.4
ENST00000368188.2 ENST00000368190.3 ENST00000368189.3 |
FCRL5
|
Fc receptor-like 5 |
| chr1_-_119682812 | 0.16 |
ENST00000537870.1
|
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr7_-_56160625 | 0.16 |
ENST00000446428.1
ENST00000432123.1 ENST00000452681.2 ENST00000537360.1 |
PHKG1
|
phosphorylase kinase, gamma 1 (muscle) |
| chr11_+_27076764 | 0.15 |
ENST00000525090.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr1_-_25256368 | 0.15 |
ENST00000308873.6
|
RUNX3
|
runt-related transcription factor 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.6 | 5.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.6 | 1.8 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.5 | 1.6 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.5 | 2.7 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.4 | 2.7 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.3 | 1.0 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.3 | 2.6 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.3 | 3.1 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.3 | 7.4 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.3 | 0.9 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.3 | 1.5 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.2 | 2.9 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.2 | 1.9 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.2 | 0.2 | GO:0042700 | luteinizing hormone signaling pathway(GO:0042700) |
| 0.2 | 0.8 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.2 | 1.8 | GO:0001554 | luteolysis(GO:0001554) |
| 0.2 | 0.6 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.2 | 1.7 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.2 | 0.6 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.2 | 0.8 | GO:0070981 | L-asparagine biosynthetic process(GO:0070981) L-asparagine metabolic process(GO:0070982) |
| 0.2 | 0.5 | GO:0042097 | negative regulation of tolerance induction(GO:0002644) interleukin-4 biosynthetic process(GO:0042097) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.2 | 1.4 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 0.5 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.1 | 1.5 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 0.7 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.1 | 1.3 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.1 | 1.0 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 | 0.8 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.1 | 0.3 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 0.3 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.1 | 0.6 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.6 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 0.6 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.1 | 0.7 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.3 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 | 1.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.6 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.1 | 0.5 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.1 | 0.6 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 0.5 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.1 | 0.3 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 0.5 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.2 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.3 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.1 | 0.7 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.3 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 0.3 | GO:0002911 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 | 0.2 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.2 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.1 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.0 | 0.9 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.1 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 1.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.6 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.6 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.3 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 1.0 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.2 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.5 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.5 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.0 | 0.2 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.4 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 2.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 1.0 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.5 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.2 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.1 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 0.2 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.3 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.6 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.2 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.5 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.5 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 0.5 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.1 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.6 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.2 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.4 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.2 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.0 | 0.4 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 5.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 0.5 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 0.8 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) protease inhibitor complex(GO:0097179) |
| 0.1 | 1.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.6 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.1 | 0.5 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.9 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 6.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.9 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 1.7 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.5 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.5 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.5 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.2 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 1.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.6 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 1.0 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 2.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.2 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.3 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 2.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.0 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 1.7 | GO:0016363 | nuclear matrix(GO:0016363) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.7 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.5 | 7.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.3 | 1.4 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.3 | 1.5 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.2 | 0.6 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.2 | 0.6 | GO:0052895 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 0.2 | 0.8 | GO:0008513 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.2 | 0.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.2 | 0.8 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.2 | 1.8 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 1.5 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 1.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 5.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.7 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.5 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.6 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.2 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.1 | 0.4 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 1.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 0.7 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 1.0 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 0.5 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.7 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.6 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 1.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.5 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.6 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.8 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.6 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 1.0 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.5 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.3 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.9 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.6 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.9 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.3 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.5 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.1 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 4.1 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 2.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.0 | GO:0045518 | interleukin-22 receptor binding(GO:0045518) |
| 0.0 | 0.6 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 1.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.3 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.5 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 1.6 | GO:0001653 | peptide receptor activity(GO:0001653) G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 0.3 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 3.1 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 7.3 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 1.7 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.8 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.8 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 1.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.5 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.8 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 1.5 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 3.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.7 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 4.0 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.8 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 2.9 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.0 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 2.3 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.5 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.5 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 3.5 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.8 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |