Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for PBX2
Z-value: 0.35
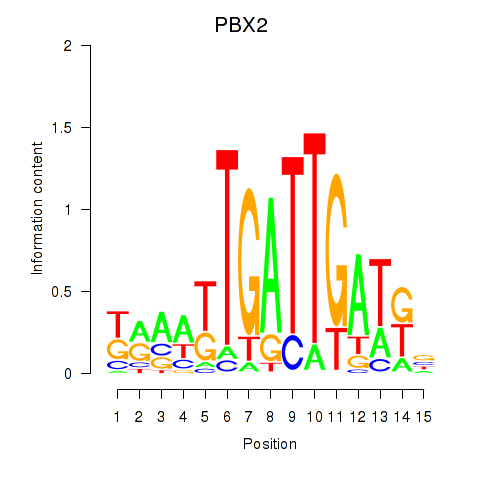
Transcription factors associated with PBX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PBX2
|
ENSG00000204304.7 | PBX homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PBX2 | hg19_v2_chr6_-_32157947_32157992 | 0.52 | 8.3e-03 | Click! |
Activity profile of PBX2 motif
Sorted Z-values of PBX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_12857015 | 1.36 |
ENST00000155926.4
|
TRIB2
|
tribbles pseudokinase 2 |
| chr2_+_12857043 | 1.34 |
ENST00000381465.2
|
TRIB2
|
tribbles pseudokinase 2 |
| chr8_-_6420930 | 0.87 |
ENST00000325203.5
|
ANGPT2
|
angiopoietin 2 |
| chr10_-_45474237 | 0.74 |
ENST00000448778.1
ENST00000298295.3 |
C10orf10
|
chromosome 10 open reading frame 10 |
| chr3_-_129158676 | 0.64 |
ENST00000393278.2
|
MBD4
|
methyl-CpG binding domain protein 4 |
| chr3_-_129158850 | 0.59 |
ENST00000503197.1
ENST00000249910.1 ENST00000429544.2 ENST00000507208.1 |
MBD4
|
methyl-CpG binding domain protein 4 |
| chr2_+_20646824 | 0.47 |
ENST00000272233.4
|
RHOB
|
ras homolog family member B |
| chr18_-_72920372 | 0.46 |
ENST00000581620.1
ENST00000582437.1 |
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2 |
| chr17_+_7758374 | 0.42 |
ENST00000301599.6
ENST00000574668.1 |
TMEM88
|
transmembrane protein 88 |
| chr11_-_2162162 | 0.36 |
ENST00000381389.1
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr11_-_104769141 | 0.33 |
ENST00000508062.1
ENST00000422698.2 |
CASP12
|
caspase 12 (gene/pseudogene) |
| chr3_+_129159039 | 0.27 |
ENST00000507564.1
ENST00000431818.2 ENST00000504021.1 ENST00000349441.2 ENST00000348417.2 ENST00000440957.2 |
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas) |
| chr3_+_129158926 | 0.27 |
ENST00000347300.2
ENST00000296266.3 |
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas) |
| chr2_+_234104079 | 0.24 |
ENST00000417661.1
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr7_-_33140498 | 0.23 |
ENST00000448915.1
|
RP9
|
retinitis pigmentosa 9 (autosomal dominant) |
| chr2_+_33661382 | 0.15 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr13_-_107220455 | 0.14 |
ENST00000400198.3
|
ARGLU1
|
arginine and glutamate rich 1 |
| chr12_-_28124903 | 0.12 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chr16_+_1128781 | 0.12 |
ENST00000293897.4
ENST00000562758.1 |
SSTR5
|
somatostatin receptor 5 |
| chr14_-_23624511 | 0.11 |
ENST00000529705.2
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr16_-_11492366 | 0.08 |
ENST00000595360.1
|
CTD-3088G3.8
|
Protein LOC388210 |
| chr10_+_124320156 | 0.07 |
ENST00000338354.3
ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr10_+_124320195 | 0.07 |
ENST00000359586.6
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr3_+_159943362 | 0.06 |
ENST00000326474.3
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr6_+_26045603 | 0.06 |
ENST00000540144.1
|
HIST1H3C
|
histone cluster 1, H3c |
| chrX_+_119737806 | 0.05 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr17_+_8191815 | 0.05 |
ENST00000226105.6
ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF
|
RAN guanine nucleotide release factor |
| chr1_+_207038699 | 0.05 |
ENST00000367098.1
|
IL20
|
interleukin 20 |
| chr14_+_58765103 | 0.05 |
ENST00000355431.3
ENST00000348476.3 ENST00000395168.3 |
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr12_-_39734783 | 0.04 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr6_+_160211481 | 0.04 |
ENST00000367034.4
|
MRPL18
|
mitochondrial ribosomal protein L18 |
| chr3_+_188889737 | 0.04 |
ENST00000345063.3
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr1_+_101702417 | 0.04 |
ENST00000305352.6
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr3_-_52864680 | 0.03 |
ENST00000406595.1
ENST00000485816.1 ENST00000434759.3 ENST00000346281.5 ENST00000266041.4 |
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chr7_+_134528635 | 0.03 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chr3_+_120626919 | 0.02 |
ENST00000273666.6
ENST00000471454.1 ENST00000472879.1 ENST00000497029.1 ENST00000492541.1 |
STXBP5L
|
syntaxin binding protein 5-like |
| chr7_-_32529973 | 0.02 |
ENST00000410044.1
ENST00000409987.1 ENST00000409782.1 ENST00000450169.2 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr2_+_234602305 | 0.02 |
ENST00000406651.1
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr10_+_47894572 | 0.02 |
ENST00000355876.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr2_+_204732666 | 0.02 |
ENST00000295854.6
ENST00000472206.1 |
CTLA4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr17_-_29151794 | 0.02 |
ENST00000324238.6
|
CRLF3
|
cytokine receptor-like factor 3 |
| chr4_-_100065419 | 0.01 |
ENST00000504125.1
ENST00000505590.1 |
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr2_+_204732487 | 0.01 |
ENST00000302823.3
|
CTLA4
|
cytotoxic T-lymphocyte-associated protein 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PBX2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.7 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.2 | 0.9 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.1 | 0.5 | GO:0060830 | intraciliary anterograde transport(GO:0035720) ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 0.2 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 1.2 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.4 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.1 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.5 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.0 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.0 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.3 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 2.7 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 0.5 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.0 | GO:0045518 | interleukin-22 receptor binding(GO:0045518) |
| 0.0 | 0.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.9 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |