Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
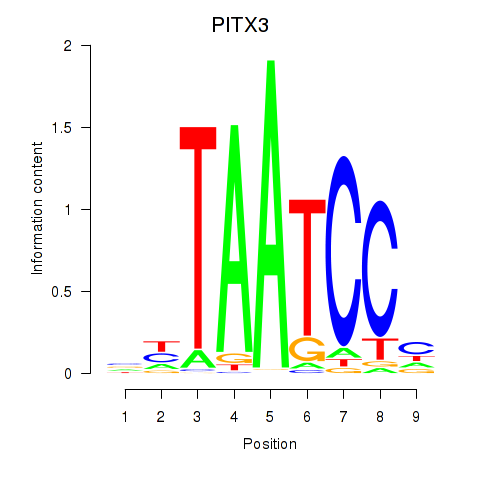
Results for PITX3
Z-value: 1.10
Transcription factors associated with PITX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PITX3
|
ENSG00000107859.5 | paired like homeodomain 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PITX3 | hg19_v2_chr10_-_104001231_104001274 | -0.44 | 2.9e-02 | Click! |
Activity profile of PITX3 motif
Sorted Z-values of PITX3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_77703115 | 3.92 |
ENST00000361092.4
ENST00000376808.4 |
NMRK1
|
nicotinamide riboside kinase 1 |
| chr1_+_35258592 | 3.82 |
ENST00000342280.4
ENST00000450137.1 |
GJA4
|
gap junction protein, alpha 4, 37kDa |
| chr4_+_126237554 | 3.19 |
ENST00000394329.3
|
FAT4
|
FAT atypical cadherin 4 |
| chr4_+_41540160 | 3.18 |
ENST00000503057.1
ENST00000511496.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr12_-_10022735 | 3.17 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr9_+_75766763 | 3.12 |
ENST00000456643.1
ENST00000415424.1 |
ANXA1
|
annexin A1 |
| chr18_+_8717369 | 2.92 |
ENST00000359865.3
ENST00000400050.3 ENST00000306285.7 |
SOGA2
|
SOGA family member 2 |
| chr2_-_188312971 | 2.77 |
ENST00000410068.1
ENST00000447403.1 ENST00000410102.1 |
CALCRL
|
calcitonin receptor-like |
| chr9_-_39288092 | 2.62 |
ENST00000323947.7
ENST00000297668.6 ENST00000377656.2 ENST00000377659.1 |
CNTNAP3
|
contactin associated protein-like 3 |
| chr4_-_186877806 | 2.53 |
ENST00000355634.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_-_100231349 | 2.41 |
ENST00000287474.5
ENST00000414213.1 |
FRRS1
|
ferric-chelate reductase 1 |
| chr4_-_90756769 | 2.40 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr9_-_77703056 | 2.39 |
ENST00000376811.1
|
NMRK1
|
nicotinamide riboside kinase 1 |
| chr4_-_186877502 | 2.31 |
ENST00000431902.1
ENST00000284776.7 ENST00000415274.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr12_-_58212487 | 2.27 |
ENST00000549994.1
|
AVIL
|
advillin |
| chr9_-_74675521 | 2.23 |
ENST00000377024.3
|
C9orf57
|
chromosome 9 open reading frame 57 |
| chr13_+_32838801 | 2.18 |
ENST00000542859.1
|
FRY
|
furry homolog (Drosophila) |
| chr21_-_46012386 | 2.16 |
ENST00000400368.1
|
KRTAP10-6
|
keratin associated protein 10-6 |
| chr4_+_140586922 | 2.04 |
ENST00000265498.1
ENST00000506797.1 |
MGST2
|
microsomal glutathione S-transferase 2 |
| chrX_-_62571187 | 2.00 |
ENST00000335144.3
|
SPIN4
|
spindlin family, member 4 |
| chr3_+_193853927 | 1.93 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr2_+_38893208 | 1.86 |
ENST00000410063.1
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chr4_-_102268628 | 1.81 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr18_+_42260861 | 1.76 |
ENST00000282030.5
|
SETBP1
|
SET binding protein 1 |
| chr2_+_120687335 | 1.75 |
ENST00000544261.1
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr19_+_2476116 | 1.74 |
ENST00000215631.4
ENST00000587345.1 |
GADD45B
|
growth arrest and DNA-damage-inducible, beta |
| chr4_-_90757364 | 1.73 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr10_+_103986085 | 1.60 |
ENST00000370005.3
|
ELOVL3
|
ELOVL fatty acid elongase 3 |
| chr12_+_51632666 | 1.59 |
ENST00000604900.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr19_+_46001697 | 1.59 |
ENST00000451287.2
ENST00000324688.4 |
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr5_-_111312622 | 1.59 |
ENST00000395634.3
|
NREP
|
neuronal regeneration related protein |
| chr1_+_163038565 | 1.59 |
ENST00000421743.2
|
RGS4
|
regulator of G-protein signaling 4 |
| chr1_+_207262881 | 1.58 |
ENST00000451804.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr21_-_31859755 | 1.57 |
ENST00000334055.3
|
KRTAP19-2
|
keratin associated protein 19-2 |
| chr12_-_42983478 | 1.55 |
ENST00000345127.3
ENST00000547113.1 |
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr16_-_23521710 | 1.55 |
ENST00000562117.1
ENST00000567468.1 ENST00000562944.1 ENST00000309859.4 |
GGA2
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 2 |
| chr6_+_132891461 | 1.51 |
ENST00000275198.1
|
TAAR6
|
trace amine associated receptor 6 |
| chr8_-_80993010 | 1.49 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr15_+_66585555 | 1.48 |
ENST00000319194.5
ENST00000525134.2 ENST00000441424.2 |
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr12_-_95009837 | 1.47 |
ENST00000551457.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr1_+_207262578 | 1.44 |
ENST00000243611.5
ENST00000367076.3 |
C4BPB
|
complement component 4 binding protein, beta |
| chr4_-_186732048 | 1.43 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_+_82266053 | 1.41 |
ENST00000370715.1
ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2
|
latrophilin 2 |
| chr20_+_33759854 | 1.41 |
ENST00000216968.4
|
PROCR
|
protein C receptor, endothelial |
| chrX_-_117107680 | 1.40 |
ENST00000447671.2
ENST00000262820.3 |
KLHL13
|
kelch-like family member 13 |
| chr7_-_15726296 | 1.39 |
ENST00000262041.5
|
MEOX2
|
mesenchyme homeobox 2 |
| chr22_+_31523734 | 1.39 |
ENST00000402238.1
ENST00000404453.1 ENST00000401755.1 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr7_+_73868439 | 1.38 |
ENST00000424337.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr5_+_95066823 | 1.38 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr3_-_149293990 | 1.35 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr11_+_5646213 | 1.34 |
ENST00000429814.2
|
TRIM34
|
tripartite motif containing 34 |
| chr4_-_140527848 | 1.34 |
ENST00000608795.1
ENST00000608958.1 |
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr3_-_158390282 | 1.33 |
ENST00000264265.3
|
LXN
|
latexin |
| chr10_-_49482907 | 1.29 |
ENST00000374201.3
ENST00000407470.4 |
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr3_-_18466026 | 1.29 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chrX_-_117107542 | 1.28 |
ENST00000371878.1
|
KLHL13
|
kelch-like family member 13 |
| chr7_+_13141097 | 1.27 |
ENST00000411542.1
|
AC011288.2
|
AC011288.2 |
| chr1_+_78470530 | 1.27 |
ENST00000370763.5
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr7_+_12727250 | 1.26 |
ENST00000404894.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr6_+_130339710 | 1.25 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr21_-_31869451 | 1.25 |
ENST00000334058.2
|
KRTAP19-4
|
keratin associated protein 19-4 |
| chr4_-_123843597 | 1.25 |
ENST00000510735.1
ENST00000304430.5 |
NUDT6
|
nudix (nucleoside diphosphate linked moiety X)-type motif 6 |
| chr7_-_72992865 | 1.21 |
ENST00000452475.1
|
TBL2
|
transducin (beta)-like 2 |
| chr4_-_102267953 | 1.21 |
ENST00000523694.2
ENST00000507176.1 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr2_-_191878162 | 1.20 |
ENST00000540176.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr17_-_14683517 | 1.20 |
ENST00000379640.1
|
AC005863.1
|
AC005863.1 |
| chr1_+_162351503 | 1.20 |
ENST00000458626.2
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr9_+_90112117 | 1.19 |
ENST00000358077.5
|
DAPK1
|
death-associated protein kinase 1 |
| chr4_-_110624564 | 1.18 |
ENST00000352981.3
ENST00000265164.2 ENST00000505486.1 |
CASP6
|
caspase 6, apoptosis-related cysteine peptidase |
| chr7_+_16793160 | 1.18 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr12_+_32655048 | 1.17 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr1_+_63249796 | 1.17 |
ENST00000443289.1
ENST00000317868.4 ENST00000371120.3 |
ATG4C
|
autophagy related 4C, cysteine peptidase |
| chr4_+_170541835 | 1.16 |
ENST00000504131.2
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr17_-_47723943 | 1.16 |
ENST00000510476.1
ENST00000503676.1 |
SPOP
|
speckle-type POZ protein |
| chr3_-_137851220 | 1.15 |
ENST00000236709.3
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr12_+_53443963 | 1.14 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr7_-_86974767 | 1.14 |
ENST00000610086.1
|
TP53TG1
|
TP53 target 1 (non-protein coding) |
| chr2_-_89247338 | 1.13 |
ENST00000496168.1
|
IGKV1-5
|
immunoglobulin kappa variable 1-5 |
| chr9_+_2015335 | 1.13 |
ENST00000349721.2
ENST00000357248.2 ENST00000450198.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr7_-_86974785 | 1.12 |
ENST00000432193.1
ENST00000421293.1 ENST00000542586.1 ENST00000359941.5 ENST00000416560.1 |
TP53TG1
|
TP53 target 1 (non-protein coding) |
| chrX_-_10851762 | 1.11 |
ENST00000380785.1
ENST00000380787.1 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr2_-_136873735 | 1.11 |
ENST00000409817.1
|
CXCR4
|
chemokine (C-X-C motif) receptor 4 |
| chr19_-_3479086 | 1.11 |
ENST00000587847.1
|
C19orf77
|
chromosome 19 open reading frame 77 |
| chr2_-_160654745 | 1.11 |
ENST00000259053.4
ENST00000429078.2 |
CD302
|
CD302 molecule |
| chr3_-_122512619 | 1.10 |
ENST00000383659.1
ENST00000306103.2 |
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1 |
| chr9_-_13279563 | 1.09 |
ENST00000541718.1
|
MPDZ
|
multiple PDZ domain protein |
| chr19_-_18995029 | 1.09 |
ENST00000596048.1
|
CERS1
|
ceramide synthase 1 |
| chr15_-_52587945 | 1.08 |
ENST00000443683.2
ENST00000558479.1 ENST00000261839.7 |
MYO5C
|
myosin VC |
| chr10_-_126716459 | 1.08 |
ENST00000309035.6
|
CTBP2
|
C-terminal binding protein 2 |
| chr9_+_75229616 | 1.08 |
ENST00000340019.3
|
TMC1
|
transmembrane channel-like 1 |
| chr1_+_66999799 | 1.08 |
ENST00000371035.3
ENST00000371036.3 ENST00000371037.4 |
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr1_-_59043166 | 1.07 |
ENST00000371225.2
|
TACSTD2
|
tumor-associated calcium signal transducer 2 |
| chr8_-_124428569 | 1.06 |
ENST00000521903.1
|
ATAD2
|
ATPase family, AAA domain containing 2 |
| chr15_-_70994612 | 1.06 |
ENST00000558758.1
ENST00000379983.2 ENST00000560441.1 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr2_+_120770581 | 1.06 |
ENST00000263713.5
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr10_-_126849588 | 1.06 |
ENST00000411419.2
|
CTBP2
|
C-terminal binding protein 2 |
| chr2_+_190722119 | 1.06 |
ENST00000452382.1
|
PMS1
|
PMS1 postmeiotic segregation increased 1 (S. cerevisiae) |
| chr11_+_63449045 | 1.06 |
ENST00000354497.4
|
RTN3
|
reticulon 3 |
| chr2_-_190627481 | 1.04 |
ENST00000264151.5
ENST00000520350.1 ENST00000521630.1 ENST00000517895.1 |
OSGEPL1
|
O-sialoglycoprotein endopeptidase-like 1 |
| chr6_+_132455118 | 1.04 |
ENST00000458028.1
|
LINC01013
|
long intergenic non-protein coding RNA 1013 |
| chr21_-_40033618 | 1.04 |
ENST00000417133.2
ENST00000398910.1 ENST00000442448.1 |
ERG
|
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr17_-_46690839 | 1.04 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chrX_-_74376108 | 1.03 |
ENST00000339447.4
ENST00000373394.3 ENST00000529949.1 ENST00000534524.1 ENST00000253577.3 |
ABCB7
|
ATP-binding cassette, sub-family B (MDR/TAP), member 7 |
| chrX_+_100333709 | 1.03 |
ENST00000372930.4
|
TMEM35
|
transmembrane protein 35 |
| chr4_-_89080003 | 1.02 |
ENST00000237612.3
|
ABCG2
|
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr21_-_35016231 | 1.02 |
ENST00000438788.1
|
CRYZL1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr12_-_2027639 | 1.01 |
ENST00000586184.1
ENST00000587995.1 ENST00000585732.1 |
CACNA2D4
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr1_+_65886326 | 1.01 |
ENST00000371059.3
ENST00000371060.3 ENST00000349533.6 ENST00000406510.3 |
LEPR
|
leptin receptor |
| chr10_-_70231639 | 1.01 |
ENST00000551118.2
ENST00000358410.3 ENST00000399180.2 ENST00000399179.2 |
DNA2
|
DNA replication helicase/nuclease 2 |
| chr12_+_52668394 | 1.01 |
ENST00000423955.2
|
KRT86
|
keratin 86 |
| chr7_+_73868220 | 1.01 |
ENST00000455841.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr2_-_11272234 | 1.00 |
ENST00000590207.1
ENST00000417697.2 ENST00000396164.1 ENST00000536743.1 ENST00000544306.1 |
AC062028.1
|
AC062028.1 |
| chr11_-_63376013 | 0.98 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr4_+_87928140 | 0.98 |
ENST00000307808.6
|
AFF1
|
AF4/FMR2 family, member 1 |
| chr2_-_74667612 | 0.97 |
ENST00000305557.5
ENST00000233330.6 |
RTKN
|
rhotekin |
| chr12_-_7848364 | 0.97 |
ENST00000329913.3
|
GDF3
|
growth differentiation factor 3 |
| chr1_+_114522049 | 0.97 |
ENST00000369551.1
ENST00000320334.4 |
OLFML3
|
olfactomedin-like 3 |
| chr7_+_73868120 | 0.96 |
ENST00000265755.3
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr11_+_118175596 | 0.96 |
ENST00000528600.1
|
CD3E
|
CD3e molecule, epsilon (CD3-TCR complex) |
| chr11_-_130184555 | 0.95 |
ENST00000525842.1
|
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr18_+_3466248 | 0.94 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr4_+_108910870 | 0.94 |
ENST00000403312.1
ENST00000603302.1 ENST00000309522.3 |
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr1_+_207262627 | 0.94 |
ENST00000391923.1
|
C4BPB
|
complement component 4 binding protein, beta |
| chr6_+_136172820 | 0.93 |
ENST00000308191.6
|
PDE7B
|
phosphodiesterase 7B |
| chr7_+_130131907 | 0.93 |
ENST00000223215.4
ENST00000437945.1 |
MEST
|
mesoderm specific transcript |
| chr4_+_159131346 | 0.93 |
ENST00000508243.1
ENST00000296529.6 |
TMEM144
|
transmembrane protein 144 |
| chr1_+_207262170 | 0.92 |
ENST00000367078.3
|
C4BPB
|
complement component 4 binding protein, beta |
| chr11_+_134123389 | 0.92 |
ENST00000281182.4
ENST00000537423.1 ENST00000543332.1 ENST00000374752.4 |
ACAD8
|
acyl-CoA dehydrogenase family, member 8 |
| chr17_-_18266765 | 0.92 |
ENST00000354098.3
|
SHMT1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr9_+_71986182 | 0.91 |
ENST00000303068.7
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr8_-_81787006 | 0.91 |
ENST00000327835.3
|
ZNF704
|
zinc finger protein 704 |
| chr9_-_13279589 | 0.91 |
ENST00000319217.7
|
MPDZ
|
multiple PDZ domain protein |
| chr8_-_77912431 | 0.91 |
ENST00000357039.4
ENST00000522527.1 |
PEX2
|
peroxisomal biogenesis factor 2 |
| chr8_-_102275131 | 0.91 |
ENST00000523121.1
|
KB-1410C5.2
|
KB-1410C5.2 |
| chr12_-_8088871 | 0.90 |
ENST00000075120.7
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr14_+_61995722 | 0.90 |
ENST00000556347.1
|
RP11-47I22.4
|
RP11-47I22.4 |
| chr7_-_150329421 | 0.89 |
ENST00000493969.1
ENST00000328902.5 |
GIMAP6
|
GTPase, IMAP family member 6 |
| chr2_-_75796837 | 0.89 |
ENST00000233712.1
|
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr1_+_24882560 | 0.89 |
ENST00000374392.2
|
NCMAP
|
noncompact myelin associated protein |
| chr14_-_22005018 | 0.88 |
ENST00000546363.1
|
SALL2
|
spalt-like transcription factor 2 |
| chrX_+_55478538 | 0.88 |
ENST00000342972.1
|
MAGEH1
|
melanoma antigen family H, 1 |
| chr15_-_34659349 | 0.87 |
ENST00000314891.6
|
LPCAT4
|
lysophosphatidylcholine acyltransferase 4 |
| chr15_-_52483566 | 0.87 |
ENST00000261837.7
|
GNB5
|
guanine nucleotide binding protein (G protein), beta 5 |
| chr8_-_93978309 | 0.87 |
ENST00000517858.1
ENST00000378861.5 |
TRIQK
|
triple QxxK/R motif containing |
| chr9_-_111775772 | 0.86 |
ENST00000325580.6
ENST00000374593.4 ENST00000374595.4 ENST00000325551.4 |
CTNNAL1
|
catenin (cadherin-associated protein), alpha-like 1 |
| chr12_-_9268707 | 0.86 |
ENST00000318602.7
|
A2M
|
alpha-2-macroglobulin |
| chr3_-_49058479 | 0.86 |
ENST00000440857.1
|
DALRD3
|
DALR anticodon binding domain containing 3 |
| chr2_+_38893047 | 0.85 |
ENST00000272252.5
|
GALM
|
galactose mutarotase (aldose 1-epimerase) |
| chrX_-_16730984 | 0.85 |
ENST00000380241.3
|
CTPS2
|
CTP synthase 2 |
| chr21_+_47706537 | 0.85 |
ENST00000397691.1
|
YBEY
|
ybeY metallopeptidase (putative) |
| chr4_+_108911036 | 0.85 |
ENST00000505878.1
|
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr12_+_109535373 | 0.85 |
ENST00000242576.2
|
UNG
|
uracil-DNA glycosylase |
| chr17_+_34087888 | 0.85 |
ENST00000586491.1
ENST00000588628.1 ENST00000285023.4 |
C17orf50
|
chromosome 17 open reading frame 50 |
| chr4_+_147096837 | 0.84 |
ENST00000296581.5
ENST00000502781.1 |
LSM6
|
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr5_-_179072047 | 0.83 |
ENST00000448248.2
|
C5orf60
|
chromosome 5 open reading frame 60 |
| chr2_-_183106641 | 0.83 |
ENST00000346717.4
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr6_-_42946947 | 0.83 |
ENST00000304611.8
|
PEX6
|
peroxisomal biogenesis factor 6 |
| chr9_+_71939488 | 0.83 |
ENST00000455972.1
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr7_+_115862858 | 0.83 |
ENST00000393481.2
|
TES
|
testis derived transcript (3 LIM domains) |
| chr17_+_56769924 | 0.82 |
ENST00000461271.1
ENST00000583539.1 ENST00000337432.4 ENST00000421782.2 |
RAD51C
|
RAD51 paralog C |
| chrX_+_107069063 | 0.82 |
ENST00000262843.6
|
MID2
|
midline 2 |
| chr19_+_21541732 | 0.82 |
ENST00000311015.3
|
ZNF738
|
zinc finger protein 738 |
| chr7_+_86975001 | 0.80 |
ENST00000412227.2
ENST00000331536.3 |
CROT
|
carnitine O-octanoyltransferase |
| chr6_-_111804905 | 0.79 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr4_+_90800656 | 0.79 |
ENST00000394980.1
|
MMRN1
|
multimerin 1 |
| chr17_+_4643337 | 0.78 |
ENST00000592813.1
|
ZMYND15
|
zinc finger, MYND-type containing 15 |
| chr3_-_65583561 | 0.78 |
ENST00000460329.2
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr15_-_55700457 | 0.78 |
ENST00000442196.3
ENST00000563171.1 ENST00000425574.3 |
CCPG1
|
cell cycle progression 1 |
| chr9_+_120466650 | 0.78 |
ENST00000355622.6
|
TLR4
|
toll-like receptor 4 |
| chr1_-_59012365 | 0.77 |
ENST00000456980.1
ENST00000482274.2 ENST00000453710.1 ENST00000419242.1 ENST00000358603.2 ENST00000371226.3 ENST00000426139.1 |
OMA1
|
OMA1 zinc metallopeptidase |
| chr14_+_64680854 | 0.77 |
ENST00000458046.2
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr14_-_106539557 | 0.77 |
ENST00000390599.2
|
IGHV1-8
|
immunoglobulin heavy variable 1-8 |
| chr1_+_63989004 | 0.77 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr22_+_23161491 | 0.77 |
ENST00000390316.2
|
IGLV3-9
|
immunoglobulin lambda variable 3-9 (gene/pseudogene) |
| chr14_-_22005062 | 0.76 |
ENST00000317492.5
|
SALL2
|
spalt-like transcription factor 2 |
| chr19_+_42381337 | 0.76 |
ENST00000597454.1
ENST00000444740.2 |
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr2_-_170430277 | 0.76 |
ENST00000438035.1
ENST00000453929.2 |
FASTKD1
|
FAST kinase domains 1 |
| chrX_-_119709637 | 0.75 |
ENST00000404115.3
|
CUL4B
|
cullin 4B |
| chr19_+_10713112 | 0.75 |
ENST00000590382.1
ENST00000407327.4 |
SLC44A2
|
solute carrier family 44 (choline transporter), member 2 |
| chr15_+_96876340 | 0.75 |
ENST00000453270.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr17_-_26127525 | 0.75 |
ENST00000313735.6
|
NOS2
|
nitric oxide synthase 2, inducible |
| chr6_-_13328564 | 0.74 |
ENST00000606530.1
ENST00000607658.1 ENST00000343141.4 ENST00000356436.4 ENST00000379300.3 ENST00000452989.1 ENST00000450347.1 ENST00000422136.1 ENST00000446018.1 ENST00000379291.1 ENST00000379307.2 ENST00000606370.1 ENST00000607230.1 |
TBC1D7
|
TBC1 domain family, member 7 |
| chr20_-_25566153 | 0.74 |
ENST00000278886.6
ENST00000422516.1 |
NINL
|
ninein-like |
| chr8_+_29953163 | 0.74 |
ENST00000518192.1
|
LEPROTL1
|
leptin receptor overlapping transcript-like 1 |
| chr13_-_31191642 | 0.74 |
ENST00000405805.1
|
HMGB1
|
high mobility group box 1 |
| chr18_+_59992514 | 0.74 |
ENST00000269485.7
|
TNFRSF11A
|
tumor necrosis factor receptor superfamily, member 11a, NFKB activator |
| chr11_-_321050 | 0.74 |
ENST00000399808.4
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr11_-_27723158 | 0.73 |
ENST00000395980.2
|
BDNF
|
brain-derived neurotrophic factor |
| chr5_+_140579162 | 0.73 |
ENST00000536699.1
ENST00000354757.3 |
PCDHB11
|
protocadherin beta 11 |
| chr8_+_74903580 | 0.73 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr9_+_36036430 | 0.72 |
ENST00000377966.3
|
RECK
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr6_+_24775153 | 0.72 |
ENST00000356509.3
ENST00000230056.3 |
GMNN
|
geminin, DNA replication inhibitor |
| chr11_+_62104897 | 0.72 |
ENST00000415229.2
ENST00000535727.1 ENST00000301776.5 |
ASRGL1
|
asparaginase like 1 |
| chr4_+_186125391 | 0.72 |
ENST00000504273.1
|
SNX25
|
sorting nexin 25 |
| chr9_+_43684902 | 0.71 |
ENST00000377564.3
ENST00000276974.6 |
CNTNAP3B
|
contactin associated protein-like 3B |
| chr19_+_21203481 | 0.71 |
ENST00000595401.1
|
ZNF430
|
zinc finger protein 430 |
| chr6_-_158589259 | 0.71 |
ENST00000367101.1
ENST00000367104.3 ENST00000367102.2 |
SERAC1
|
serine active site containing 1 |
| chr9_-_100459639 | 0.71 |
ENST00000375128.4
|
XPA
|
xeroderma pigmentosum, complementation group A |
| chr2_+_132285406 | 0.71 |
ENST00000295171.6
ENST00000409856.3 |
CCDC74A
|
coiled-coil domain containing 74A |
| chr2_-_70475701 | 0.70 |
ENST00000282574.4
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of PITX3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0014839 | myoblast migration involved in skeletal muscle regeneration(GO:0014839) |
| 1.0 | 3.0 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.8 | 4.1 | GO:0051622 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.8 | 3.2 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.7 | 2.7 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.7 | 3.3 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.7 | 2.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.6 | 5.6 | GO:1903027 | regulation of opsonization(GO:1903027) |
| 0.6 | 1.8 | GO:1904482 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.5 | 2.5 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.4 | 2.5 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.4 | 2.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.4 | 1.6 | GO:2000691 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.4 | 1.1 | GO:0036146 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.3 | 1.0 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.3 | 1.0 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.3 | 1.2 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.3 | 0.9 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.3 | 0.9 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.3 | 0.8 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.3 | 2.2 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.3 | 1.6 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.3 | 0.8 | GO:0070428 | negative regulation of interleukin-23 production(GO:0032707) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.3 | 2.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.3 | 2.8 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.2 | 0.7 | GO:0071812 | circadian temperature homeostasis(GO:0060086) regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.2 | 0.7 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.2 | 1.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.2 | 0.9 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.2 | 0.9 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.2 | 0.9 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.2 | 0.7 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.2 | 3.0 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.2 | 1.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.2 | 1.3 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 0.2 | 0.6 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.2 | 1.0 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.2 | 2.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 1.0 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.2 | 0.6 | GO:0015728 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.2 | 0.6 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.2 | 1.2 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.2 | 1.0 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.2 | 0.8 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.2 | 1.1 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.2 | 6.5 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 1.5 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.2 | 0.5 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.2 | 1.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.2 | 1.1 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.2 | 1.6 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 0.5 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.2 | 1.6 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.2 | 0.7 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.2 | 0.9 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.2 | 0.9 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.2 | 0.7 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.2 | 1.7 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 1.2 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.2 | 0.7 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.2 | 1.9 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 0.5 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.2 | 0.3 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.2 | 1.3 | GO:0075044 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.2 | 6.2 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.2 | 0.9 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.2 | 0.6 | GO:0072554 | blood vessel lumenization(GO:0072554) |
| 0.2 | 1.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.2 | 0.6 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.2 | 0.9 | GO:0001757 | somite specification(GO:0001757) |
| 0.2 | 0.5 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.1 | 0.4 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 1.2 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.7 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 0.3 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.1 | 0.4 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.1 | 0.6 | GO:1901091 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.1 | 0.6 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.6 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 1.3 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.1 | 0.8 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.1 | 0.8 | GO:0061518 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 0.4 | GO:0046707 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.1 | 1.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 1.1 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.1 | 1.6 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.7 | GO:0021938 | ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.1 | 0.5 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 0.8 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 0.6 | GO:1901678 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) iron coordination entity transport(GO:1901678) |
| 0.1 | 1.1 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.1 | 1.0 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.1 | 0.1 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 0.4 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.1 | 1.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 1.3 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.1 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 0.7 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.6 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.1 | 1.4 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 0.7 | GO:0034182 | regulation of maintenance of sister chromatid cohesion(GO:0034091) regulation of maintenance of mitotic sister chromatid cohesion(GO:0034182) |
| 0.1 | 0.5 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.6 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.1 | 0.4 | GO:1905167 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.1 | 1.9 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 0.3 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.1 | 0.2 | GO:0002605 | negative regulation of dendritic cell antigen processing and presentation(GO:0002605) |
| 0.1 | 0.9 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 2.1 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.1 | 0.5 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 | 1.2 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.1 | 0.3 | GO:0042040 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.1 | 0.7 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 1.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.5 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 0.4 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.3 | GO:2001162 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.1 | 0.3 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.8 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.1 | 0.5 | GO:0045337 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.1 | 0.6 | GO:0050893 | sensory processing(GO:0050893) |
| 0.1 | 1.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.3 | GO:0007497 | posterior midgut development(GO:0007497) endothelin receptor signaling pathway(GO:0086100) |
| 0.1 | 0.3 | GO:0060545 | regulation of nucleoside transport(GO:0032242) positive regulation of necroptotic process(GO:0060545) |
| 0.1 | 0.3 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.1 | 1.5 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 1.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.9 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 1.4 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 0.4 | GO:1903566 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.4 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 0.6 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 0.6 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 0.3 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.1 | 2.7 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 0.4 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.1 | 0.4 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.1 | 1.0 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.1 | 0.5 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.1 | 0.3 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.1 | 0.3 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 0.3 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.1 | 0.6 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 0.8 | GO:0032310 | prostaglandin secretion(GO:0032310) |
| 0.1 | 3.5 | GO:0048265 | response to pain(GO:0048265) |
| 0.1 | 0.8 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.2 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.1 | 1.1 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 | 0.6 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.1 | 1.3 | GO:0060004 | reflex(GO:0060004) |
| 0.1 | 1.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.2 | GO:0048867 | ganglion mother cell fate determination(GO:0007402) stem cell fate determination(GO:0048867) |
| 0.1 | 0.5 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.1 | 0.4 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 0.3 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.1 | 0.2 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.1 | 0.1 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.1 | 0.5 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 | 0.2 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.1 | 0.2 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 0.7 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.1 | 0.1 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.1 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.2 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 0.6 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 0.2 | GO:0003064 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.4 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.1 | 0.6 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.1 | 0.2 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.1 | 0.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.2 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 1.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.5 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.1 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.1 | 0.4 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.2 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 | 0.3 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 0.2 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.1 | 0.3 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.1 | 1.6 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.3 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 0.3 | GO:1904386 | response to vitamin K(GO:0032571) response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.1 | 0.3 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.1 | 0.2 | GO:0000494 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.1 | 0.4 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 0.7 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.1 | 0.3 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 0.9 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.1 | 0.2 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.1 | 0.3 | GO:0019075 | virus maturation(GO:0019075) |
| 0.1 | 0.2 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 1.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 2.2 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 0.2 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.1 | 0.3 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.1 | 0.3 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.1 | 0.8 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.2 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.1 | 0.4 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.1 | 0.8 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.1 | 1.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.2 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.1 | 0.2 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.1 | 0.2 | GO:0046104 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.2 | GO:0002784 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 0.8 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 0.4 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.3 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.1 | 1.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 1.0 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 2.5 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.1 | 0.2 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.1 | 0.4 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.2 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.1 | 0.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 0.6 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.4 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.1 | 1.0 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.1 | 0.2 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.1 | 0.2 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 0.4 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.8 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 1.2 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.2 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.3 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.1 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.0 | 0.4 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.4 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.3 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 0.7 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.7 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.9 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.1 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.0 | 0.6 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.4 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.4 | GO:0071352 | cellular response to interleukin-2(GO:0071352) |
| 0.0 | 1.2 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.2 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.2 | GO:2001153 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.0 | 1.9 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.6 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) negative regulation of growth of symbiont in host(GO:0044130) |
| 0.0 | 0.3 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.3 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.2 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.0 | 1.3 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.2 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.9 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.5 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.1 | GO:0015993 | molecular hydrogen transport(GO:0015993) |
| 0.0 | 1.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.5 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.5 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 0.1 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 7.9 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.2 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.3 | GO:0098704 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.1 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.3 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.3 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.5 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.3 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.3 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 0.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.4 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.2 | GO:1902172 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.0 | 1.0 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.8 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 0.4 | GO:0071415 | cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.0 | 0.2 | GO:2001184 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.0 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.0 | 1.1 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.2 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.5 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 1.2 | GO:1901661 | quinone metabolic process(GO:1901661) |
| 0.0 | 0.2 | GO:0038030 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.3 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 1.7 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.7 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.8 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.2 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.3 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.0 | 0.2 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.2 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 1.8 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.0 | 0.3 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.2 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.1 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) |
| 0.0 | 0.3 | GO:0009304 | tRNA transcription(GO:0009304) |
| 0.0 | 0.2 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 0.5 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.0 | 0.5 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.2 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.4 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.5 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 1.0 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.5 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.4 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.0 | 0.2 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.0 | 0.7 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.2 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.2 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.3 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.2 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.3 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.2 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.2 | GO:0072506 | cellular phosphate ion homeostasis(GO:0030643) phosphate ion homeostasis(GO:0055062) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) trivalent inorganic anion homeostasis(GO:0072506) |
| 0.0 | 0.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.3 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.1 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.1 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.1 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.1 | GO:0045423 | regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.0 | 0.9 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.5 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.3 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.4 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.3 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.4 | GO:0034776 | response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.2 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.1 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.0 | 0.3 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.1 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:0072108 | positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0072108) |
| 0.0 | 0.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 1.2 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 1.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.0 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.0 | 0.9 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.7 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.2 | GO:0006577 | amino-acid betaine metabolic process(GO:0006577) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.4 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.1 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.7 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.4 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.3 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.1 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 0.2 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.0 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.0 | 0.1 | GO:0052314 | response to pyrethroid(GO:0046684) phytoalexin metabolic process(GO:0052314) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.2 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:2000910 | negative regulation of cytokine secretion involved in immune response(GO:0002740) negative regulation of cholesterol import(GO:0060621) negative regulation of sterol import(GO:2000910) |
| 0.0 | 0.2 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.0 | 0.1 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.3 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.4 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.2 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.9 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 1.6 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.6 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.2 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.1 | GO:2000849 | glucocorticoid secretion(GO:0035933) regulation of glucocorticoid secretion(GO:2000849) |
| 0.0 | 0.1 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.1 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.4 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.4 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.4 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.4 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.2 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.0 | 0.3 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 | 0.3 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.3 | GO:0006525 | arginine metabolic process(GO:0006525) |
| 0.0 | 0.1 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.5 | GO:0002209 | behavioral fear response(GO:0001662) behavioral defense response(GO:0002209) |
| 0.0 | 0.1 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.6 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.2 | GO:0090520 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.2 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.0 | 0.2 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.3 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.0 | 0.1 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.0 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 0.0 | 0.0 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.2 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 1.7 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.2 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.2 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.3 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.8 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.0 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:0061740 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) |
| 0.0 | 0.2 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.1 | GO:1904778 | protein localization to cell cortex(GO:0072697) regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.0 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.2 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0014029 | neural crest formation(GO:0014029) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.8 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.3 | 3.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.3 | 1.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.3 | 0.9 | GO:0036028 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.2 | 0.9 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.2 | 0.4 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.2 | 0.7 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.2 | 0.6 | GO:0000939 | nuclear MIS12/MIND complex(GO:0000818) condensed chromosome inner kinetochore(GO:0000939) |
| 0.2 | 0.4 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.2 | 0.6 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.2 | 1.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 1.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.2 | 1.4 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.2 | 2.1 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.2 | 0.5 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.2 | 0.9 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.2 | 2.0 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 1.0 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 3.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 5.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.6 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 2.6 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 1.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 2.9 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 1.4 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 0.6 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 0.4 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.5 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.4 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 0.6 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.1 | 0.9 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.3 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.7 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 0.7 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.5 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 1.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.8 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.4 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 1.7 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.3 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.1 | 0.5 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 0.3 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.1 | 1.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.9 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 0.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.3 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 0.3 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 1.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.7 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 0.8 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 2.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.4 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.9 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.2 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.1 | 0.4 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.1 | 0.3 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 0.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 0.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.3 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.1 | 1.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 0.6 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 1.7 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 0.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 1.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.3 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 0.2 | GO:0097196 | Shu complex(GO:0097196) |
| 0.1 | 0.8 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 0.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.5 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.1 | 0.5 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 0.2 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.1 | 0.9 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.1 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 3.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 1.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 1.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.9 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.3 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.3 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.2 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.2 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.0 | 2.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.4 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 2.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.7 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.1 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.2 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.6 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.8 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.6 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.7 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.4 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.4 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 2.0 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 1.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.7 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 3.2 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 2.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.7 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 1.1 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.0 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 4.0 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 2.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 2.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0097059 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.0 | GO:0042571 | immunoglobulin complex(GO:0019814) immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.2 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 4.0 | GO:0031674 | I band(GO:0031674) |
| 0.0 | 0.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) 3M complex(GO:1990393) |
| 0.0 | 0.0 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 1.5 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 1.5 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.2 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.6 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0031262 | condensed nuclear chromosome outer kinetochore(GO:0000942) Ndc80 complex(GO:0031262) |
| 0.0 | 0.0 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 0.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.3 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.4 | GO:0098553 | integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 1.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.6 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.8 | 4.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.7 | 2.8 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.7 | 2.0 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.5 | 3.6 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.5 | 2.8 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.4 | 2.5 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.4 | 1.9 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.4 | 1.8 | GO:0070905 | serine binding(GO:0070905) |
| 0.3 | 1.0 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.3 | 0.6 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.3 | 2.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.3 | 3.0 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.3 | 0.9 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.3 | 0.9 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.3 | 0.9 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.3 | 0.8 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.3 | 1.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 7.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 1.0 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.2 | 0.7 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.2 | 0.7 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.2 | 0.9 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.2 | 0.7 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.2 | 2.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.2 | 1.0 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.2 | 0.8 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.2 | 1.4 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 0.6 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.2 | 0.8 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.2 | 0.7 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.2 | 1.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 1.6 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 0.5 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.2 | 0.7 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.2 | 1.5 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.2 | 1.9 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 1.4 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.2 | 0.6 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 0.5 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.1 | 2.7 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 2.6 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 0.7 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.4 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.1 | 1.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.3 | GO:0070404 | NADH binding(GO:0070404) |
| 0.1 | 1.0 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.1 | 0.5 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 1.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 1.8 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.7 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 1.0 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.4 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.1 | 1.5 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.1 | 1.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 0.5 | GO:1904492 | Ac-Asp-Glu binding(GO:1904492) tetrahydrofolyl-poly(glutamate) polymer binding(GO:1904493) |
| 0.1 | 0.3 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.1 | 0.5 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.1 | 0.6 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.1 | 0.5 | GO:0019144 | ADP-sugar diphosphatase activity(GO:0019144) |
| 0.1 | 0.6 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.1 | 0.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 2.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.4 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 0.7 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 0.4 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.3 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.1 | 0.5 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.1 | 0.5 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.3 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 0.4 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.1 | 0.8 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 0.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 1.0 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.1 | 0.3 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.8 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.6 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.6 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 1.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.5 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.1 | 0.4 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 1.3 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.3 | GO:0034188 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.6 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.3 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 0.6 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 0.2 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.1 | 1.0 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.4 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 0.2 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.1 | 2.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.7 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 1.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.2 | GO:0018636 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) |
| 0.1 | 0.5 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.1 | 1.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 1.0 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 1.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.2 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.1 | 0.2 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 1.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.6 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.1 | 0.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.3 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.1 | 0.4 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.4 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 0.4 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.8 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.3 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 0.2 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 0.1 | 2.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.3 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.1 | 1.2 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.2 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.1 | 0.5 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.1 | 0.4 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 2.0 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.2 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.1 | 0.2 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.1 | 0.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.2 | GO:0016160 | amylase activity(GO:0016160) |
| 0.1 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.5 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.3 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.1 | 0.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 1.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.3 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.1 | 0.2 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.1 | 0.2 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.1 | 0.2 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.1 | 1.1 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 0.3 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.7 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.3 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.7 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.3 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.0 | 1.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.2 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.3 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 1.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.7 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.3 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.5 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.7 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 1.0 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.6 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.6 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.3 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 1.6 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.2 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 0.1 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.0 | 1.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 1.5 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) |
| 0.0 | 0.3 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.3 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 1.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 1.0 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.3 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.0 | 0.9 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.2 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.1 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 1.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.5 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.6 | GO:1900750 | oligopeptide binding(GO:1900750) |
| 0.0 | 0.4 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 1.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 1.1 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.0 | 1.2 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 1.6 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 3.0 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.1 | GO:0005503 | all-trans retinal binding(GO:0005503) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.4 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.2 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.2 | GO:0004673 | phosphorelay sensor kinase activity(GO:0000155) protein histidine kinase activity(GO:0004673) C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.2 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.9 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.6 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 1.2 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.5 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.7 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.2 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.7 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 1.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.0 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.0 | 1.0 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.5 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 1.3 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.8 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 0.1 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 0.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.3 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.4 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 1.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.8 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.8 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.9 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.3 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.0 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.7 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.0 | GO:0005499 | vitamin D binding(GO:0005499) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 4.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 4.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 1.0 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 4.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.5 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.9 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 2.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 2.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 2.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.5 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 5.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.4 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.5 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.2 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.2 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.5 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.2 | PID ENDOTHELIN PATHWAY | Endothelins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 3.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 2.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 1.8 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 1.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 0.8 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.1 | 0.6 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 1.7 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.5 | REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | Genes involved in Signaling by the B Cell Receptor (BCR) |
| 0.1 | 1.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 0.6 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 1.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 2.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 2.1 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 1.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.3 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.3 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 1.3 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 1.1 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.9 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 2.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.9 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.7 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 2.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 5.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 3.3 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.8 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 2.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.4 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 1.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 0.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.2 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.3 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |