Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
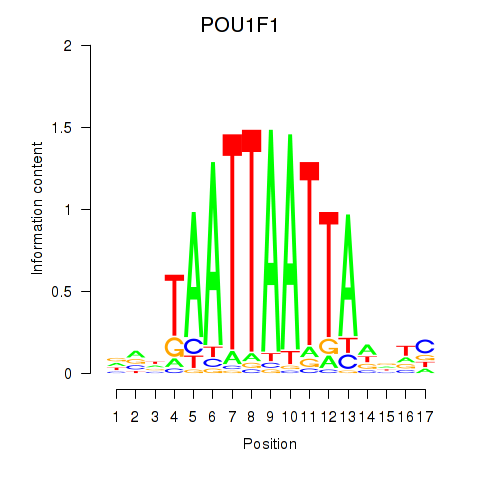
Results for POU1F1
Z-value: 0.49
Transcription factors associated with POU1F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU1F1
|
ENSG00000064835.6 | POU class 1 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU1F1 | hg19_v2_chr3_-_87325612_87325654 | 0.11 | 6.0e-01 | Click! |
Activity profile of POU1F1 motif
Sorted Z-values of POU1F1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_67418047 | 1.38 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr2_+_161993465 | 1.26 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr11_-_102651343 | 1.18 |
ENST00000279441.4
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 (stromelysin 2) |
| chr2_+_152214098 | 1.06 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr2_+_161993412 | 1.01 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr10_+_91152303 | 0.97 |
ENST00000371804.3
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr5_+_55149150 | 0.83 |
ENST00000297015.3
|
IL31RA
|
interleukin 31 receptor A |
| chr9_+_12693336 | 0.82 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr10_-_49860525 | 0.58 |
ENST00000435790.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr7_-_14026063 | 0.54 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr11_+_35201826 | 0.53 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr22_+_23487513 | 0.53 |
ENST00000263116.2
ENST00000341989.4 |
RAB36
|
RAB36, member RAS oncogene family |
| chr19_+_54466179 | 0.53 |
ENST00000270458.2
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8 |
| chr17_-_5321549 | 0.49 |
ENST00000572809.1
|
NUP88
|
nucleoporin 88kDa |
| chr6_+_114178512 | 0.49 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr2_+_103035102 | 0.49 |
ENST00000264260.2
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr3_-_191000172 | 0.49 |
ENST00000427544.2
|
UTS2B
|
urotensin 2B |
| chr4_-_120243545 | 0.47 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr11_+_5710919 | 0.40 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr2_-_231989808 | 0.38 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr17_+_29664830 | 0.37 |
ENST00000444181.2
ENST00000417592.2 |
NF1
|
neurofibromin 1 |
| chr9_-_85882145 | 0.37 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr10_-_14050522 | 0.37 |
ENST00000342409.2
|
FRMD4A
|
FERM domain containing 4A |
| chr17_+_76494911 | 0.37 |
ENST00000598378.1
|
DNAH17-AS1
|
DNAH17 antisense RNA 1 |
| chr2_-_220264703 | 0.35 |
ENST00000519905.1
ENST00000523282.1 ENST00000434339.1 ENST00000457935.1 |
DNPEP
|
aspartyl aminopeptidase |
| chr1_+_8378140 | 0.33 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr12_-_11150474 | 0.33 |
ENST00000538986.1
|
TAS2R20
|
taste receptor, type 2, member 20 |
| chr4_+_74301880 | 0.32 |
ENST00000395792.2
ENST00000226359.2 |
AFP
|
alpha-fetoprotein |
| chr4_+_71263599 | 0.32 |
ENST00000399575.2
|
PROL1
|
proline rich, lacrimal 1 |
| chr1_+_87012753 | 0.31 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chr7_-_139763521 | 0.31 |
ENST00000263549.3
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr7_+_134528635 | 0.30 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chr3_-_108248169 | 0.28 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr17_-_39165366 | 0.28 |
ENST00000391588.1
|
KRTAP3-1
|
keratin associated protein 3-1 |
| chr5_-_88119580 | 0.28 |
ENST00000539796.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr19_+_572543 | 0.27 |
ENST00000333511.3
ENST00000573216.1 ENST00000353555.4 |
BSG
|
basigin (Ok blood group) |
| chr14_-_101295407 | 0.26 |
ENST00000596284.1
|
AL117190.2
|
AL117190.2 |
| chr3_-_27498235 | 0.26 |
ENST00000295736.5
ENST00000428386.1 ENST00000428179.1 |
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr4_+_96012614 | 0.26 |
ENST00000264568.4
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr5_+_111755280 | 0.26 |
ENST00000600409.1
|
EPB41L4A-AS2
|
EPB41L4A antisense RNA 2 (head to head) |
| chr3_+_52245458 | 0.26 |
ENST00000459884.1
|
ALAS1
|
aminolevulinate, delta-, synthase 1 |
| chr7_+_13141097 | 0.26 |
ENST00000411542.1
|
AC011288.2
|
AC011288.2 |
| chr5_+_173472607 | 0.25 |
ENST00000303177.3
ENST00000519867.1 |
NSG2
|
Neuron-specific protein family member 2 |
| chr9_-_33473882 | 0.25 |
ENST00000455041.2
ENST00000353159.2 ENST00000297990.4 ENST00000379471.2 |
NOL6
|
nucleolar protein 6 (RNA-associated) |
| chr10_-_50396425 | 0.25 |
ENST00000374148.1
|
C10orf128
|
chromosome 10 open reading frame 128 |
| chr6_+_26440700 | 0.24 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr16_-_28634874 | 0.24 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr4_+_86525299 | 0.23 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr16_-_70835034 | 0.23 |
ENST00000261776.5
|
VAC14
|
Vac14 homolog (S. cerevisiae) |
| chr6_+_26365443 | 0.23 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr10_-_27529486 | 0.23 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr5_-_93077293 | 0.22 |
ENST00000510627.4
|
POU5F2
|
POU domain class 5, transcription factor 2 |
| chr3_+_159943362 | 0.22 |
ENST00000326474.3
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr8_+_30244580 | 0.22 |
ENST00000523115.1
ENST00000519647.1 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr1_-_247921982 | 0.22 |
ENST00000408896.2
|
OR1C1
|
olfactory receptor, family 1, subfamily C, member 1 |
| chr6_-_89927151 | 0.22 |
ENST00000454853.2
|
GABRR1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chrX_-_11308598 | 0.21 |
ENST00000380717.3
|
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr10_+_53806501 | 0.21 |
ENST00000373975.2
|
PRKG1
|
protein kinase, cGMP-dependent, type I |
| chr1_-_67266939 | 0.20 |
ENST00000304526.2
|
INSL5
|
insulin-like 5 |
| chr4_+_186990298 | 0.20 |
ENST00000296795.3
ENST00000513189.1 |
TLR3
|
toll-like receptor 3 |
| chr15_-_75748115 | 0.20 |
ENST00000360439.4
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr17_-_2996290 | 0.20 |
ENST00000331459.1
|
OR1D2
|
olfactory receptor, family 1, subfamily D, member 2 |
| chr4_-_122085469 | 0.19 |
ENST00000057513.3
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr3_-_108476231 | 0.19 |
ENST00000295755.6
|
RETNLB
|
resistin like beta |
| chr19_+_19144384 | 0.19 |
ENST00000392335.2
ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr7_-_87342564 | 0.18 |
ENST00000265724.3
ENST00000416177.1 |
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chrX_-_77225135 | 0.18 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chrX_+_84258832 | 0.17 |
ENST00000373173.2
|
APOOL
|
apolipoprotein O-like |
| chr3_-_190167571 | 0.17 |
ENST00000354905.2
|
TMEM207
|
transmembrane protein 207 |
| chr15_+_84115868 | 0.17 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr19_-_22193731 | 0.17 |
ENST00000601773.1
ENST00000397126.4 ENST00000601993.1 ENST00000599916.1 |
ZNF208
|
zinc finger protein 208 |
| chr22_-_43010928 | 0.16 |
ENST00000348657.2
ENST00000252115.5 |
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr15_+_75970672 | 0.16 |
ENST00000435356.1
|
AC105020.1
|
Uncharacterized protein; cDNA FLJ12988 fis, clone NT2RP3000080 |
| chr9_-_95056010 | 0.16 |
ENST00000443024.2
|
IARS
|
isoleucyl-tRNA synthetase |
| chr19_-_56826157 | 0.16 |
ENST00000592509.1
ENST00000592679.1 ENST00000588442.1 ENST00000593106.1 ENST00000587492.1 ENST00000254165.3 |
ZSCAN5A
|
zinc finger and SCAN domain containing 5A |
| chr4_-_159956333 | 0.16 |
ENST00000434826.2
|
C4orf45
|
chromosome 4 open reading frame 45 |
| chr12_+_12510352 | 0.16 |
ENST00000298571.6
|
LOH12CR1
|
loss of heterozygosity, 12, chromosomal region 1 |
| chr4_+_175839506 | 0.16 |
ENST00000505141.1
ENST00000359240.3 ENST00000445694.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr4_+_3344141 | 0.15 |
ENST00000306648.7
|
RGS12
|
regulator of G-protein signaling 12 |
| chr3_+_160939050 | 0.15 |
ENST00000493066.1
ENST00000351193.2 ENST00000472947.1 ENST00000463518.1 |
NMD3
|
NMD3 ribosome export adaptor |
| chr1_+_111682058 | 0.15 |
ENST00000545121.1
|
CEPT1
|
choline/ethanolamine phosphotransferase 1 |
| chr1_+_117963209 | 0.15 |
ENST00000449370.2
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr12_-_95510743 | 0.15 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chrY_-_6740649 | 0.15 |
ENST00000383036.1
ENST00000383037.4 |
AMELY
|
amelogenin, Y-linked |
| chr12_-_86650045 | 0.15 |
ENST00000604798.1
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr21_-_27423339 | 0.15 |
ENST00000415997.1
|
APP
|
amyloid beta (A4) precursor protein |
| chr5_+_140762268 | 0.15 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr8_-_38008783 | 0.14 |
ENST00000276449.4
|
STAR
|
steroidogenic acute regulatory protein |
| chr4_-_103746683 | 0.14 |
ENST00000504211.1
ENST00000508476.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr4_+_175839551 | 0.14 |
ENST00000404450.4
ENST00000514159.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr11_-_104827425 | 0.14 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr9_-_215744 | 0.14 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr8_+_36641842 | 0.14 |
ENST00000523973.1
ENST00000399881.3 |
KCNU1
|
potassium channel, subfamily U, member 1 |
| chr11_+_2405833 | 0.14 |
ENST00000527343.1
ENST00000464784.2 |
CD81
|
CD81 molecule |
| chr19_+_52873166 | 0.14 |
ENST00000424032.2
ENST00000600321.1 ENST00000344085.5 ENST00000597976.1 ENST00000422689.2 |
ZNF880
|
zinc finger protein 880 |
| chr17_-_64225508 | 0.14 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr4_-_122854612 | 0.13 |
ENST00000264811.5
|
TRPC3
|
transient receptor potential cation channel, subfamily C, member 3 |
| chr18_+_29027696 | 0.13 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr11_-_107729887 | 0.13 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr21_-_15583165 | 0.13 |
ENST00000536861.1
|
LIPI
|
lipase, member I |
| chr1_+_234765057 | 0.13 |
ENST00000429269.1
|
LINC00184
|
long intergenic non-protein coding RNA 184 |
| chr15_+_64680003 | 0.13 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr9_+_111624577 | 0.13 |
ENST00000333999.3
|
ACTL7A
|
actin-like 7A |
| chr6_-_136788001 | 0.13 |
ENST00000544465.1
|
MAP7
|
microtubule-associated protein 7 |
| chr1_-_13390765 | 0.13 |
ENST00000357367.2
|
PRAMEF8
|
PRAME family member 8 |
| chr12_-_52761262 | 0.12 |
ENST00000257901.3
|
KRT85
|
keratin 85 |
| chr7_-_111032971 | 0.12 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr18_+_45778672 | 0.12 |
ENST00000600091.1
|
AC091150.1
|
HCG1818186; Uncharacterized protein |
| chrX_+_70503433 | 0.12 |
ENST00000276079.8
ENST00000373856.3 ENST00000373841.1 ENST00000420903.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr20_+_59654146 | 0.12 |
ENST00000441660.1
|
RP5-827L5.1
|
RP5-827L5.1 |
| chr18_+_21269404 | 0.12 |
ENST00000313654.9
|
LAMA3
|
laminin, alpha 3 |
| chr7_+_129984630 | 0.12 |
ENST00000355388.3
ENST00000497503.1 ENST00000463587.1 ENST00000461828.1 ENST00000494311.1 ENST00000466363.2 ENST00000485477.1 ENST00000431780.2 ENST00000474905.1 |
CPA5
|
carboxypeptidase A5 |
| chr17_+_6918354 | 0.12 |
ENST00000552775.1
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr6_-_134861089 | 0.12 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr3_+_130650738 | 0.12 |
ENST00000504612.1
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr6_+_29555683 | 0.12 |
ENST00000383640.2
|
OR2H2
|
olfactory receptor, family 2, subfamily H, member 2 |
| chr6_-_152639479 | 0.12 |
ENST00000356820.4
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr5_+_156696362 | 0.12 |
ENST00000377576.3
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr9_-_95055956 | 0.12 |
ENST00000375629.3
ENST00000447699.2 ENST00000375643.3 ENST00000395554.3 |
IARS
|
isoleucyl-tRNA synthetase |
| chr1_-_183560011 | 0.12 |
ENST00000367536.1
|
NCF2
|
neutrophil cytosolic factor 2 |
| chr20_-_43589109 | 0.12 |
ENST00000372813.3
|
TOMM34
|
translocase of outer mitochondrial membrane 34 |
| chr10_+_94352956 | 0.12 |
ENST00000260731.3
|
KIF11
|
kinesin family member 11 |
| chr1_-_183559693 | 0.12 |
ENST00000367535.3
ENST00000413720.1 ENST00000418089.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr4_+_151503077 | 0.11 |
ENST00000317605.4
|
MAB21L2
|
mab-21-like 2 (C. elegans) |
| chr1_+_112938803 | 0.11 |
ENST00000271277.6
|
CTTNBP2NL
|
CTTNBP2 N-terminal like |
| chr2_+_223162866 | 0.11 |
ENST00000295226.1
|
CCDC140
|
coiled-coil domain containing 140 |
| chr1_-_182921119 | 0.11 |
ENST00000423786.1
|
SHCBP1L
|
SHC SH2-domain binding protein 1-like |
| chr7_+_99425633 | 0.11 |
ENST00000354829.2
ENST00000421837.2 ENST00000417625.1 ENST00000342499.4 ENST00000444905.1 ENST00000415413.1 ENST00000312017.5 ENST00000222382.5 |
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43 |
| chrX_+_83116142 | 0.11 |
ENST00000329312.4
|
CYLC1
|
cylicin, basic protein of sperm head cytoskeleton 1 |
| chr18_+_21269556 | 0.11 |
ENST00000399516.3
|
LAMA3
|
laminin, alpha 3 |
| chr12_+_93096759 | 0.11 |
ENST00000544406.2
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr6_-_109702885 | 0.11 |
ENST00000504373.1
|
CD164
|
CD164 molecule, sialomucin |
| chr8_-_59412717 | 0.10 |
ENST00000301645.3
|
CYP7A1
|
cytochrome P450, family 7, subfamily A, polypeptide 1 |
| chr12_+_69753448 | 0.10 |
ENST00000247843.2
ENST00000548020.1 ENST00000549685.1 ENST00000552955.1 |
YEATS4
|
YEATS domain containing 4 |
| chr11_-_58612168 | 0.10 |
ENST00000287275.1
|
GLYATL2
|
glycine-N-acyltransferase-like 2 |
| chr12_-_10959892 | 0.10 |
ENST00000240615.2
|
TAS2R8
|
taste receptor, type 2, member 8 |
| chr20_+_5987890 | 0.10 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr5_+_119867159 | 0.10 |
ENST00000505123.1
|
PRR16
|
proline rich 16 |
| chrX_-_32173579 | 0.09 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr1_+_152943122 | 0.09 |
ENST00000328051.2
|
SPRR4
|
small proline-rich protein 4 |
| chr11_-_22647350 | 0.09 |
ENST00000327470.3
|
FANCF
|
Fanconi anemia, complementation group F |
| chr2_-_190044480 | 0.09 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr10_+_97759848 | 0.09 |
ENST00000424464.1
ENST00000410012.2 ENST00000344386.3 |
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chr4_+_119809984 | 0.09 |
ENST00000307142.4
ENST00000448416.2 ENST00000429713.2 |
SYNPO2
|
synaptopodin 2 |
| chr14_-_25078864 | 0.09 |
ENST00000216338.4
ENST00000557220.2 ENST00000382548.4 |
GZMH
|
granzyme H (cathepsin G-like 2, protein h-CCPX) |
| chr2_+_58655461 | 0.09 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr5_+_60933634 | 0.09 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr1_+_119957554 | 0.09 |
ENST00000543831.1
ENST00000433745.1 ENST00000369416.3 |
HSD3B2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr17_+_1944790 | 0.09 |
ENST00000575162.1
|
DPH1
|
diphthamide biosynthesis 1 |
| chr17_+_6918093 | 0.09 |
ENST00000439424.2
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr12_+_93096619 | 0.09 |
ENST00000397833.3
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr2_-_99871570 | 0.09 |
ENST00000333017.2
ENST00000409679.1 ENST00000423306.1 |
LYG2
|
lysozyme G-like 2 |
| chr12_-_25150373 | 0.09 |
ENST00000549828.1
|
C12orf77
|
chromosome 12 open reading frame 77 |
| chr4_-_138453559 | 0.09 |
ENST00000511115.1
|
PCDH18
|
protocadherin 18 |
| chr7_-_141957847 | 0.08 |
ENST00000552471.1
ENST00000547058.2 |
PRSS58
|
protease, serine, 58 |
| chr5_+_140593509 | 0.08 |
ENST00000341948.4
|
PCDHB13
|
protocadherin beta 13 |
| chr1_-_53608289 | 0.08 |
ENST00000371491.4
|
SLC1A7
|
solute carrier family 1 (glutamate transporter), member 7 |
| chr10_-_52645379 | 0.08 |
ENST00000395489.2
|
A1CF
|
APOBEC1 complementation factor |
| chr11_-_13517565 | 0.08 |
ENST00000282091.1
ENST00000529816.1 |
PTH
|
parathyroid hormone |
| chr3_+_132843652 | 0.08 |
ENST00000508711.1
|
TMEM108
|
transmembrane protein 108 |
| chr10_+_62538089 | 0.08 |
ENST00000519078.2
ENST00000395284.3 ENST00000316629.4 |
CDK1
|
cyclin-dependent kinase 1 |
| chr12_+_25205666 | 0.08 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr12_+_26348429 | 0.08 |
ENST00000242729.2
|
SSPN
|
sarcospan |
| chr6_-_49604545 | 0.08 |
ENST00000371175.4
ENST00000229810.7 |
RHAG
|
Rh-associated glycoprotein |
| chr12_-_42631529 | 0.08 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr5_-_36301984 | 0.07 |
ENST00000502994.1
ENST00000515759.1 ENST00000296604.3 |
RANBP3L
|
RAN binding protein 3-like |
| chr1_+_192127578 | 0.07 |
ENST00000367460.3
|
RGS18
|
regulator of G-protein signaling 18 |
| chr21_-_15918618 | 0.07 |
ENST00000400564.1
ENST00000400566.1 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr21_-_43735628 | 0.07 |
ENST00000291525.10
ENST00000518498.1 |
TFF3
|
trefoil factor 3 (intestinal) |
| chr20_+_5986756 | 0.07 |
ENST00000452938.1
|
CRLS1
|
cardiolipin synthase 1 |
| chr13_-_70682590 | 0.07 |
ENST00000377844.4
|
KLHL1
|
kelch-like family member 1 |
| chr3_-_160823040 | 0.07 |
ENST00000484127.1
ENST00000492353.1 ENST00000473142.1 ENST00000468268.1 ENST00000460353.1 ENST00000320474.4 ENST00000392781.2 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr12_+_26348246 | 0.07 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr7_+_134671234 | 0.07 |
ENST00000436302.2
ENST00000359383.3 ENST00000458078.1 ENST00000435976.2 ENST00000455283.2 |
AGBL3
|
ATP/GTP binding protein-like 3 |
| chr10_+_695888 | 0.07 |
ENST00000441152.2
|
PRR26
|
proline rich 26 |
| chr9_-_28670283 | 0.07 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr4_+_37455536 | 0.07 |
ENST00000381980.4
ENST00000508175.1 |
C4orf19
|
chromosome 4 open reading frame 19 |
| chr2_-_169887827 | 0.07 |
ENST00000263817.6
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chrX_-_106243451 | 0.07 |
ENST00000355610.4
ENST00000535534.1 |
MORC4
|
MORC family CW-type zinc finger 4 |
| chr1_+_149754227 | 0.07 |
ENST00000444948.1
ENST00000369168.4 |
FCGR1A
|
Fc fragment of IgG, high affinity Ia, receptor (CD64) |
| chr6_-_49712147 | 0.07 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr4_-_46911248 | 0.07 |
ENST00000355591.3
ENST00000505102.1 |
COX7B2
|
cytochrome c oxidase subunit VIIb2 |
| chr11_-_26593677 | 0.06 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr4_+_119810134 | 0.06 |
ENST00000434046.2
|
SYNPO2
|
synaptopodin 2 |
| chr17_-_72772462 | 0.06 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr10_-_28623368 | 0.06 |
ENST00000441595.2
|
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr1_-_154580616 | 0.06 |
ENST00000368474.4
|
ADAR
|
adenosine deaminase, RNA-specific |
| chrX_-_102531717 | 0.06 |
ENST00000372680.1
|
TCEAL5
|
transcription elongation factor A (SII)-like 5 |
| chr6_-_49712123 | 0.06 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr1_+_158323755 | 0.06 |
ENST00000368157.1
ENST00000368156.1 ENST00000368155.3 ENST00000368154.1 ENST00000368160.3 ENST00000368161.3 |
CD1E
|
CD1e molecule |
| chr16_+_28565230 | 0.06 |
ENST00000317058.3
|
CCDC101
|
coiled-coil domain containing 101 |
| chr6_+_131958436 | 0.06 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chrX_-_102943022 | 0.06 |
ENST00000433176.2
|
MORF4L2
|
mortality factor 4 like 2 |
| chr20_+_30697298 | 0.06 |
ENST00000398022.2
|
TM9SF4
|
transmembrane 9 superfamily protein member 4 |
| chr14_-_106668095 | 0.06 |
ENST00000390606.2
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr5_+_115177178 | 0.06 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr1_-_197036364 | 0.06 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr13_-_81801115 | 0.06 |
ENST00000567258.1
|
LINC00564
|
long intergenic non-protein coding RNA 564 |
| chr2_-_26700900 | 0.06 |
ENST00000338581.6
ENST00000339598.3 ENST00000402415.3 |
OTOF
|
otoferlin |
| chr9_-_21351377 | 0.06 |
ENST00000380210.1
|
IFNA6
|
interferon, alpha 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU1F1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.3 | 1.0 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.3 | 1.4 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 0.4 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 | 0.3 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 | 0.4 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 0.8 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.1 | 0.6 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 0.3 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.5 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.3 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 1.5 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.8 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.2 | GO:0034343 | microglial cell activation involved in immune response(GO:0002282) type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.2 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.1 | GO:0070859 | dibenzo-p-dioxin metabolic process(GO:0018894) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.0 | 0.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.0 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.1 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 0.2 | GO:1904386 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.2 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.0 | 0.1 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.3 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.0 | 0.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.1 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.2 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.5 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.0 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.0 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.0 | GO:0042138 | meiotic DNA double-strand break processing(GO:0000706) meiotic DNA double-strand break formation(GO:0042138) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.2 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 1.2 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.0 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.0 | 0.1 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.1 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.2 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 0.5 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 0.5 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.8 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0033150 | cytoskeletal calyx(GO:0033150) acrosomal matrix(GO:0043159) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.1 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 1.2 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.3 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 1.4 | GO:0043657 | host(GO:0018995) host cell(GO:0043657) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.3 | 1.4 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.3 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 0.2 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 0.2 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 1.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.0 | 0.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.2 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.4 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.0 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.0 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 1.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.0 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 1.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 1.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.5 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |