Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
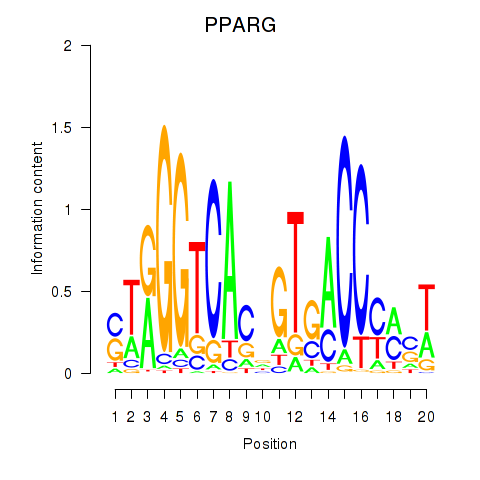
Results for PPARG
Z-value: 0.41
Transcription factors associated with PPARG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PPARG
|
ENSG00000132170.15 | peroxisome proliferator activated receptor gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PPARG | hg19_v2_chr3_+_12330560_12330579, hg19_v2_chr3_+_12329397_12329433, hg19_v2_chr3_+_12329358_12329393, hg19_v2_chr3_+_12392971_12392983 | -0.34 | 1.0e-01 | Click! |
Activity profile of PPARG motif
Sorted Z-values of PPARG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_49661037 | 1.12 |
ENST00000427978.2
|
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr19_+_49661079 | 0.76 |
ENST00000355712.5
|
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr19_+_49660997 | 0.68 |
ENST00000598691.1
ENST00000252826.5 |
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr4_+_74735102 | 0.65 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr5_+_131409476 | 0.36 |
ENST00000296871.2
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chr19_-_1592828 | 0.30 |
ENST00000592012.1
|
MBD3
|
methyl-CpG binding domain protein 3 |
| chr8_-_101348408 | 0.25 |
ENST00000519527.1
ENST00000522369.1 |
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr7_+_155090271 | 0.24 |
ENST00000476756.1
|
INSIG1
|
insulin induced gene 1 |
| chr1_+_203274639 | 0.24 |
ENST00000290551.4
|
BTG2
|
BTG family, member 2 |
| chr11_-_65667997 | 0.23 |
ENST00000312562.2
ENST00000534222.1 |
FOSL1
|
FOS-like antigen 1 |
| chr19_-_49864746 | 0.23 |
ENST00000598810.1
|
TEAD2
|
TEA domain family member 2 |
| chr22_+_31488433 | 0.21 |
ENST00000455608.1
|
SMTN
|
smoothelin |
| chr19_-_39368887 | 0.20 |
ENST00000340740.3
ENST00000591812.1 |
RINL
|
Ras and Rab interactor-like |
| chr11_+_61248583 | 0.19 |
ENST00000432063.2
ENST00000338608.2 |
PPP1R32
|
protein phosphatase 1, regulatory subunit 32 |
| chr22_+_31277661 | 0.19 |
ENST00000454145.1
ENST00000453621.1 ENST00000431368.1 ENST00000535268.1 |
OSBP2
|
oxysterol binding protein 2 |
| chr14_+_105992906 | 0.19 |
ENST00000392519.2
|
TMEM121
|
transmembrane protein 121 |
| chr14_+_105212297 | 0.18 |
ENST00000556623.1
ENST00000555674.1 |
ADSSL1
|
adenylosuccinate synthase like 1 |
| chr17_+_61571746 | 0.18 |
ENST00000579409.1
|
ACE
|
angiotensin I converting enzyme |
| chr1_-_27286897 | 0.17 |
ENST00000320567.5
|
C1orf172
|
chromosome 1 open reading frame 172 |
| chr19_+_507299 | 0.17 |
ENST00000359315.5
|
TPGS1
|
tubulin polyglutamylase complex subunit 1 |
| chr19_+_42388437 | 0.17 |
ENST00000378152.4
ENST00000337665.4 |
ARHGEF1
|
Rho guanine nucleotide exchange factor (GEF) 1 |
| chr7_+_142919130 | 0.17 |
ENST00000408947.3
|
TAS2R40
|
taste receptor, type 2, member 40 |
| chr15_-_74658519 | 0.17 |
ENST00000450547.1
ENST00000358632.4 |
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr15_-_74658493 | 0.16 |
ENST00000419019.2
ENST00000569662.1 |
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr22_+_22676808 | 0.16 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr1_-_16482554 | 0.15 |
ENST00000358432.5
|
EPHA2
|
EPH receptor A2 |
| chr2_-_74619152 | 0.15 |
ENST00000440727.1
ENST00000409240.1 |
DCTN1
|
dynactin 1 |
| chr20_+_37434329 | 0.15 |
ENST00000299824.1
ENST00000373331.2 |
PPP1R16B
|
protein phosphatase 1, regulatory subunit 16B |
| chr6_-_4079334 | 0.15 |
ENST00000492651.1
ENST00000498677.1 ENST00000274673.3 |
FAM217A
|
family with sequence similarity 217, member A |
| chr2_-_74618964 | 0.15 |
ENST00000417090.1
ENST00000409868.1 |
DCTN1
|
dynactin 1 |
| chr22_-_46659219 | 0.14 |
ENST00000253255.5
|
PKDREJ
|
polycystin (PKD) family receptor for egg jelly |
| chr15_-_82939157 | 0.14 |
ENST00000559949.1
|
RP13-996F3.5
|
golgin A6 family-like 18 |
| chr17_+_48172639 | 0.14 |
ENST00000503176.1
ENST00000503614.1 |
PDK2
|
pyruvate dehydrogenase kinase, isozyme 2 |
| chr17_+_31318886 | 0.14 |
ENST00000269053.3
ENST00000394638.1 |
SPACA3
|
sperm acrosome associated 3 |
| chr4_-_10023095 | 0.14 |
ENST00000264784.3
|
SLC2A9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr19_-_44031375 | 0.14 |
ENST00000292147.2
|
ETHE1
|
ethylmalonic encephalopathy 1 |
| chr16_+_29817399 | 0.14 |
ENST00000545521.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr9_-_117150243 | 0.13 |
ENST00000374088.3
|
AKNA
|
AT-hook transcription factor |
| chr11_-_1593150 | 0.13 |
ENST00000397374.3
|
DUSP8
|
dual specificity phosphatase 8 |
| chr19_-_1592652 | 0.13 |
ENST00000156825.1
ENST00000434436.3 |
MBD3
|
methyl-CpG binding domain protein 3 |
| chr11_-_65686496 | 0.13 |
ENST00000449692.3
|
C11orf68
|
chromosome 11 open reading frame 68 |
| chr19_-_46405861 | 0.13 |
ENST00000322217.5
|
MYPOP
|
Myb-related transcription factor, partner of profilin |
| chr20_+_816695 | 0.13 |
ENST00000246100.3
|
FAM110A
|
family with sequence similarity 110, member A |
| chr8_+_145203548 | 0.13 |
ENST00000534366.1
|
MROH1
|
maestro heat-like repeat family member 1 |
| chr19_+_42772659 | 0.13 |
ENST00000572681.2
|
CIC
|
capicua transcriptional repressor |
| chr2_+_201994208 | 0.12 |
ENST00000440180.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr19_+_50016411 | 0.12 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr5_+_38445641 | 0.12 |
ENST00000397210.3
ENST00000506135.1 ENST00000508131.1 |
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains |
| chr19_-_42498231 | 0.12 |
ENST00000602133.1
|
ATP1A3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr19_+_56152262 | 0.12 |
ENST00000325333.5
ENST00000590190.1 |
ZNF580
|
zinc finger protein 580 |
| chr16_+_8768422 | 0.12 |
ENST00000268251.8
ENST00000564714.1 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr19_-_55690758 | 0.12 |
ENST00000590851.1
|
SYT5
|
synaptotagmin V |
| chrY_+_9236030 | 0.12 |
ENST00000457222.2
ENST00000440483.1 ENST00000424594.1 ENST00000427871.2 ENST00000423647.2 |
TSPY3
TSPY1
|
testis specific protein, Y-linked 3 testis specific protein, Y-linked 1 |
| chr3_-_133748913 | 0.11 |
ENST00000310926.4
|
SLCO2A1
|
solute carrier organic anion transporter family, member 2A1 |
| chr12_-_125348329 | 0.11 |
ENST00000546215.1
ENST00000415380.2 ENST00000261693.6 ENST00000376788.1 ENST00000545493.1 |
SCARB1
|
scavenger receptor class B, member 1 |
| chr1_+_158323244 | 0.11 |
ENST00000434258.1
|
CD1E
|
CD1e molecule |
| chr19_+_4279282 | 0.11 |
ENST00000599689.1
|
SHD
|
Src homology 2 domain containing transforming protein D |
| chr5_+_32710736 | 0.11 |
ENST00000415685.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chrX_+_153770421 | 0.11 |
ENST00000369609.5
ENST00000369607.1 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr2_+_120187465 | 0.11 |
ENST00000409826.1
ENST00000417645.1 |
TMEM37
|
transmembrane protein 37 |
| chr1_+_207070775 | 0.11 |
ENST00000391929.3
ENST00000294984.2 ENST00000367093.3 |
IL24
|
interleukin 24 |
| chr11_-_65686586 | 0.11 |
ENST00000438576.2
|
C11orf68
|
chromosome 11 open reading frame 68 |
| chr7_-_38407770 | 0.11 |
ENST00000390348.2
|
TRGV1
|
T cell receptor gamma variable 1 (non-functional) |
| chr15_-_74753443 | 0.11 |
ENST00000567435.1
ENST00000564488.1 ENST00000565130.1 ENST00000563081.1 ENST00000565335.1 ENST00000395081.2 ENST00000361351.4 |
UBL7
|
ubiquitin-like 7 (bone marrow stromal cell-derived) |
| chr10_+_114135952 | 0.11 |
ENST00000356116.1
ENST00000433418.1 ENST00000354273.4 |
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr9_-_130524695 | 0.11 |
ENST00000440630.1
ENST00000429553.1 |
SH2D3C
|
SH2 domain containing 3C |
| chr19_-_10445399 | 0.10 |
ENST00000592945.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr1_+_16083154 | 0.10 |
ENST00000375771.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr19_+_50016610 | 0.10 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr11_-_64901978 | 0.10 |
ENST00000294256.8
ENST00000377190.3 |
SYVN1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr11_-_64901900 | 0.10 |
ENST00000526060.1
ENST00000307289.6 ENST00000528487.1 |
SYVN1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr11_+_76494253 | 0.10 |
ENST00000333090.4
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr12_-_53601000 | 0.10 |
ENST00000338737.4
ENST00000549086.2 |
ITGB7
|
integrin, beta 7 |
| chr19_+_35609380 | 0.10 |
ENST00000604621.1
|
FXYD3
|
FXYD domain containing ion transport regulator 3 |
| chr8_+_39442097 | 0.09 |
ENST00000265707.5
ENST00000379866.1 ENST00000520772.1 ENST00000541111.1 |
ADAM18
|
ADAM metallopeptidase domain 18 |
| chr4_+_926171 | 0.09 |
ENST00000507319.1
ENST00000264771.4 |
TMEM175
|
transmembrane protein 175 |
| chr19_+_4472230 | 0.09 |
ENST00000301284.4
ENST00000586684.1 |
HDGFRP2
|
Hepatoma-derived growth factor-related protein 2 |
| chr11_-_6440283 | 0.09 |
ENST00000299402.6
ENST00000609360.1 ENST00000389906.2 ENST00000532020.2 |
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr19_+_56717283 | 0.09 |
ENST00000376267.1
|
ZSCAN5C
|
zinc finger and SCAN domain containing 5C |
| chr22_-_50312052 | 0.09 |
ENST00000330817.6
|
ALG12
|
ALG12, alpha-1,6-mannosyltransferase |
| chr2_+_201994042 | 0.09 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr20_+_814377 | 0.09 |
ENST00000304189.2
ENST00000381939.1 |
FAM110A
|
family with sequence similarity 110, member A |
| chr11_+_46740730 | 0.09 |
ENST00000311907.5
ENST00000530231.1 ENST00000442468.1 |
F2
|
coagulation factor II (thrombin) |
| chr20_+_814349 | 0.09 |
ENST00000381941.3
|
FAM110A
|
family with sequence similarity 110, member A |
| chr17_-_27045405 | 0.09 |
ENST00000430132.2
|
RAB34
|
RAB34, member RAS oncogene family |
| chr7_-_100425112 | 0.08 |
ENST00000358173.3
|
EPHB4
|
EPH receptor B4 |
| chr1_-_111970353 | 0.08 |
ENST00000369732.3
|
OVGP1
|
oviductal glycoprotein 1, 120kDa |
| chr5_-_180076613 | 0.08 |
ENST00000261937.6
ENST00000393347.3 |
FLT4
|
fms-related tyrosine kinase 4 |
| chrY_+_9304564 | 0.08 |
ENST00000451548.1
|
TSPY1
|
testis specific protein, Y-linked 1 |
| chr8_-_25902876 | 0.08 |
ENST00000520164.1
|
EBF2
|
early B-cell factor 2 |
| chr11_-_64052111 | 0.08 |
ENST00000394532.3
ENST00000394531.3 ENST00000309032.3 |
BAD
|
BCL2-associated agonist of cell death |
| chr12_-_125348448 | 0.08 |
ENST00000339570.5
|
SCARB1
|
scavenger receptor class B, member 1 |
| chr17_-_62097904 | 0.08 |
ENST00000583366.1
|
ICAM2
|
intercellular adhesion molecule 2 |
| chr4_-_860950 | 0.08 |
ENST00000511980.1
ENST00000510799.1 |
GAK
|
cyclin G associated kinase |
| chr9_-_138591341 | 0.08 |
ENST00000298466.5
ENST00000425225.1 |
SOHLH1
|
spermatogenesis and oogenesis specific basic helix-loop-helix 1 |
| chr1_+_3816936 | 0.08 |
ENST00000413332.1
ENST00000442673.1 ENST00000439488.1 |
RP13-15E13.1
|
long intergenic non-protein coding RNA 1134 |
| chr1_-_47407111 | 0.08 |
ENST00000371904.4
|
CYP4A11
|
cytochrome P450, family 4, subfamily A, polypeptide 11 |
| chr17_-_74287333 | 0.08 |
ENST00000447564.2
|
QRICH2
|
glutamine rich 2 |
| chr1_+_217804661 | 0.08 |
ENST00000366933.4
|
SPATA17
|
spermatogenesis associated 17 |
| chr16_+_29817841 | 0.08 |
ENST00000322945.6
ENST00000562337.1 ENST00000566906.2 ENST00000563402.1 ENST00000219782.6 |
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr2_+_28853443 | 0.08 |
ENST00000436775.1
|
PLB1
|
phospholipase B1 |
| chrY_+_9175073 | 0.08 |
ENST00000426950.2
ENST00000383008.1 |
TSPY4
|
testis specific protein, Y-linked 4 |
| chr19_+_18451391 | 0.08 |
ENST00000269919.6
ENST00000604499.2 ENST00000595066.1 ENST00000252813.5 |
PGPEP1
|
pyroglutamyl-peptidase I |
| chr20_-_56286479 | 0.07 |
ENST00000265626.4
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr7_-_38370536 | 0.07 |
ENST00000390343.2
|
TRGV8
|
T cell receptor gamma variable 8 |
| chr11_-_67210930 | 0.07 |
ENST00000453768.2
ENST00000545016.1 ENST00000341356.5 |
CORO1B
|
coronin, actin binding protein, 1B |
| chr17_-_27044810 | 0.07 |
ENST00000395242.2
|
RAB34
|
RAB34, member RAS oncogene family |
| chr11_-_82612727 | 0.07 |
ENST00000531128.1
ENST00000535099.1 ENST00000527444.1 |
PRCP
|
prolylcarboxypeptidase (angiotensinase C) |
| chr19_-_13213662 | 0.07 |
ENST00000264824.4
|
LYL1
|
lymphoblastic leukemia derived sequence 1 |
| chr7_-_19184929 | 0.07 |
ENST00000275461.3
|
FERD3L
|
Fer3-like bHLH transcription factor |
| chr6_+_33378738 | 0.07 |
ENST00000374512.3
ENST00000374516.3 |
PHF1
|
PHD finger protein 1 |
| chrY_+_6114264 | 0.07 |
ENST00000320701.4
ENST00000383042.1 |
TSPY2
|
testis specific protein, Y-linked 2 |
| chr19_-_14228541 | 0.07 |
ENST00000590853.1
ENST00000308677.4 |
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha |
| chr17_-_42092313 | 0.07 |
ENST00000587529.1
ENST00000206380.3 ENST00000542039.1 |
TMEM101
|
transmembrane protein 101 |
| chr5_-_180076580 | 0.07 |
ENST00000502649.1
|
FLT4
|
fms-related tyrosine kinase 4 |
| chr17_+_80709932 | 0.07 |
ENST00000355528.4
ENST00000397466.2 ENST00000539345.2 |
TBCD
|
tubulin folding cofactor D |
| chr6_-_117086873 | 0.07 |
ENST00000368557.4
|
FAM162B
|
family with sequence similarity 162, member B |
| chr6_-_49712147 | 0.07 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chrY_+_9195406 | 0.07 |
ENST00000287721.9
ENST00000383005.2 ENST00000383000.1 ENST00000330628.9 ENST00000537415.1 |
TSPY8
|
testis specific protein, Y-linked 8 |
| chr13_+_107029084 | 0.07 |
ENST00000444865.1
|
LINC00460
|
long intergenic non-protein coding RNA 460 |
| chr12_+_66582919 | 0.07 |
ENST00000545837.1
ENST00000457197.2 |
IRAK3
|
interleukin-1 receptor-associated kinase 3 |
| chr21_-_33984456 | 0.07 |
ENST00000431216.1
ENST00000553001.1 ENST00000440966.1 |
AP000275.65
C21orf59
|
Uncharacterized protein chromosome 21 open reading frame 59 |
| chrX_+_153775821 | 0.07 |
ENST00000263518.6
ENST00000470142.1 ENST00000393549.2 ENST00000455588.2 ENST00000369602.3 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr15_+_41186609 | 0.07 |
ENST00000220509.5
|
VPS18
|
vacuolar protein sorting 18 homolog (S. cerevisiae) |
| chr16_-_11492366 | 0.07 |
ENST00000595360.1
|
CTD-3088G3.8
|
Protein LOC388210 |
| chr7_-_150498426 | 0.07 |
ENST00000447204.2
|
TMEM176B
|
transmembrane protein 176B |
| chr20_+_44563267 | 0.07 |
ENST00000372409.3
|
PCIF1
|
PDX1 C-terminal inhibiting factor 1 |
| chr2_+_220306745 | 0.06 |
ENST00000431523.1
ENST00000396698.1 ENST00000396695.2 |
SPEG
|
SPEG complex locus |
| chr9_-_35112376 | 0.06 |
ENST00000488109.2
|
FAM214B
|
family with sequence similarity 214, member B |
| chr1_-_223853348 | 0.06 |
ENST00000366872.5
|
CAPN8
|
calpain 8 |
| chr1_-_161147275 | 0.06 |
ENST00000319769.5
ENST00000367998.1 |
B4GALT3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
| chr4_-_21546272 | 0.06 |
ENST00000509207.1
|
KCNIP4
|
Kv channel interacting protein 4 |
| chr12_+_6494285 | 0.06 |
ENST00000541102.1
|
LTBR
|
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr19_-_36399149 | 0.06 |
ENST00000585901.2
ENST00000544690.2 ENST00000424586.3 ENST00000262629.4 ENST00000589517.1 |
TYROBP
|
TYRO protein tyrosine kinase binding protein |
| chr17_-_62097927 | 0.06 |
ENST00000578313.1
ENST00000584084.1 ENST00000579788.1 ENST00000579687.1 ENST00000578379.1 ENST00000578892.1 ENST00000412356.1 ENST00000418105.1 |
ICAM2
|
intercellular adhesion molecule 2 |
| chr2_+_219283815 | 0.06 |
ENST00000248444.5
ENST00000454069.1 ENST00000392114.2 |
VIL1
|
villin 1 |
| chrY_+_9324922 | 0.06 |
ENST00000440215.2
ENST00000446779.2 |
TSPY6P
|
testis specific protein, Y-linked 6, pseudogene |
| chr1_-_11115877 | 0.06 |
ENST00000490101.1
|
SRM
|
spermidine synthase |
| chr7_-_75443118 | 0.06 |
ENST00000222902.2
|
CCL24
|
chemokine (C-C motif) ligand 24 |
| chr9_+_84603687 | 0.06 |
ENST00000344803.2
|
SPATA31D1
|
SPATA31 subfamily D, member 1 |
| chr17_-_27044760 | 0.06 |
ENST00000395243.3
|
RAB34
|
RAB34, member RAS oncogene family |
| chr1_-_40137710 | 0.06 |
ENST00000235628.1
|
NT5C1A
|
5'-nucleotidase, cytosolic IA |
| chr16_+_20912382 | 0.06 |
ENST00000396052.2
|
LYRM1
|
LYR motif containing 1 |
| chr9_-_117150303 | 0.06 |
ENST00000312033.3
|
AKNA
|
AT-hook transcription factor |
| chr9_+_136243117 | 0.06 |
ENST00000426926.2
ENST00000371957.3 |
C9orf96
|
chromosome 9 open reading frame 96 |
| chr9_+_139847347 | 0.06 |
ENST00000371632.3
|
LCN12
|
lipocalin 12 |
| chr4_+_41992489 | 0.06 |
ENST00000264451.7
|
SLC30A9
|
solute carrier family 30 (zinc transporter), member 9 |
| chr3_+_100120441 | 0.06 |
ENST00000489752.1
|
LNP1
|
leukemia NUP98 fusion partner 1 |
| chr22_+_19929130 | 0.06 |
ENST00000361682.6
ENST00000403184.1 ENST00000403710.1 ENST00000407537.1 |
COMT
|
catechol-O-methyltransferase |
| chr1_+_158323486 | 0.06 |
ENST00000444681.2
ENST00000368167.3 |
CD1E
|
CD1e molecule |
| chr9_+_95997205 | 0.06 |
ENST00000411624.1
|
WNK2
|
WNK lysine deficient protein kinase 2 |
| chr9_-_124976185 | 0.06 |
ENST00000464484.2
|
LHX6
|
LIM homeobox 6 |
| chr17_-_62658186 | 0.06 |
ENST00000262435.9
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr19_+_58180303 | 0.06 |
ENST00000318203.5
|
ZSCAN4
|
zinc finger and SCAN domain containing 4 |
| chr21_-_33985127 | 0.06 |
ENST00000290155.3
|
C21orf59
|
chromosome 21 open reading frame 59 |
| chr17_-_77023723 | 0.06 |
ENST00000577521.1
|
C1QTNF1-AS1
|
C1QTNF1 antisense RNA 1 |
| chr8_-_13134045 | 0.06 |
ENST00000512044.2
|
DLC1
|
deleted in liver cancer 1 |
| chr16_+_20912075 | 0.06 |
ENST00000219168.4
|
LYRM1
|
LYR motif containing 1 |
| chr11_+_60609537 | 0.06 |
ENST00000227520.5
|
CCDC86
|
coiled-coil domain containing 86 |
| chr22_+_23241661 | 0.06 |
ENST00000390322.2
|
IGLJ2
|
immunoglobulin lambda joining 2 |
| chr9_+_86237963 | 0.06 |
ENST00000277124.8
|
IDNK
|
idnK, gluconokinase homolog (E. coli) |
| chr6_+_97010424 | 0.05 |
ENST00000541107.1
ENST00000450218.1 ENST00000326771.2 |
FHL5
|
four and a half LIM domains 5 |
| chr1_+_156096336 | 0.05 |
ENST00000504687.1
ENST00000473598.2 |
LMNA
|
lamin A/C |
| chr14_-_106453155 | 0.05 |
ENST00000390594.2
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr9_+_136243264 | 0.05 |
ENST00000371955.1
|
C9orf96
|
chromosome 9 open reading frame 96 |
| chr6_+_41196077 | 0.05 |
ENST00000448827.2
|
TREML4
|
triggering receptor expressed on myeloid cells-like 4 |
| chr6_+_41196052 | 0.05 |
ENST00000341495.2
|
TREML4
|
triggering receptor expressed on myeloid cells-like 4 |
| chr17_+_55162453 | 0.05 |
ENST00000575322.1
ENST00000337714.3 ENST00000314126.3 |
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr11_+_117070904 | 0.05 |
ENST00000529792.1
|
TAGLN
|
transgelin |
| chr17_-_27045427 | 0.05 |
ENST00000301043.6
ENST00000412625.1 |
RAB34
|
RAB34, member RAS oncogene family |
| chr11_+_126225789 | 0.05 |
ENST00000530591.1
ENST00000534083.1 |
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr20_+_62152077 | 0.05 |
ENST00000370179.3
ENST00000370177.1 |
PPDPF
|
pancreatic progenitor cell differentiation and proliferation factor |
| chr22_+_29702996 | 0.05 |
ENST00000406549.3
ENST00000360113.2 ENST00000341313.6 ENST00000403764.1 ENST00000471961.1 ENST00000407854.1 |
GAS2L1
|
growth arrest-specific 2 like 1 |
| chr17_-_39942940 | 0.05 |
ENST00000310706.5
ENST00000393931.3 ENST00000424457.1 ENST00000591690.1 |
JUP
|
junction plakoglobin |
| chr19_+_45349630 | 0.05 |
ENST00000252483.5
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B) |
| chr9_-_124976154 | 0.05 |
ENST00000482062.1
|
LHX6
|
LIM homeobox 6 |
| chr14_+_23340822 | 0.05 |
ENST00000359591.4
|
LRP10
|
low density lipoprotein receptor-related protein 10 |
| chr20_-_33999766 | 0.05 |
ENST00000349714.5
ENST00000438533.1 ENST00000359226.2 ENST00000374384.2 ENST00000374377.5 ENST00000407996.2 ENST00000424405.1 ENST00000542501.1 ENST00000397554.1 ENST00000540457.1 ENST00000374380.2 ENST00000374385.5 |
UQCC1
|
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr10_-_28623368 | 0.05 |
ENST00000441595.2
|
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr21_-_33984888 | 0.05 |
ENST00000382549.4
ENST00000540881.1 |
C21orf59
|
chromosome 21 open reading frame 59 |
| chr8_+_9009202 | 0.05 |
ENST00000518496.1
|
RP11-10A14.4
|
Uncharacterized protein |
| chr19_+_18451439 | 0.05 |
ENST00000597431.2
|
PGPEP1
|
pyroglutamyl-peptidase I |
| chr19_+_56154913 | 0.05 |
ENST00000270451.5
ENST00000588537.1 |
ZNF581
|
zinc finger protein 581 |
| chr17_-_27044903 | 0.05 |
ENST00000395245.3
|
RAB34
|
RAB34, member RAS oncogene family |
| chr17_-_27045165 | 0.05 |
ENST00000436730.3
ENST00000450529.1 ENST00000583538.1 ENST00000419712.3 ENST00000580843.2 ENST00000582934.1 ENST00000415040.2 ENST00000353676.5 ENST00000453384.3 ENST00000447716.1 |
RAB34
|
RAB34, member RAS oncogene family |
| chr19_-_2783363 | 0.05 |
ENST00000221566.2
|
SGTA
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr2_-_32490801 | 0.05 |
ENST00000360906.5
ENST00000342905.6 |
NLRC4
|
NLR family, CARD domain containing 4 |
| chr8_-_145652336 | 0.05 |
ENST00000529182.1
ENST00000526054.1 |
VPS28
|
vacuolar protein sorting 28 homolog (S. cerevisiae) |
| chr4_+_57371509 | 0.05 |
ENST00000360096.2
|
ARL9
|
ADP-ribosylation factor-like 9 |
| chr19_+_4343584 | 0.05 |
ENST00000596722.1
|
MPND
|
MPN domain containing |
| chr19_-_44031341 | 0.05 |
ENST00000600651.1
|
ETHE1
|
ethylmalonic encephalopathy 1 |
| chr11_-_118134997 | 0.05 |
ENST00000278937.2
|
MPZL2
|
myelin protein zero-like 2 |
| chr2_-_32490859 | 0.05 |
ENST00000404025.2
|
NLRC4
|
NLR family, CARD domain containing 4 |
| chr15_+_40674963 | 0.05 |
ENST00000448395.2
|
KNSTRN
|
kinetochore-localized astrin/SPAG5 binding protein |
| chr3_+_119217376 | 0.05 |
ENST00000494664.1
ENST00000493694.1 |
TIMMDC1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chr12_-_96390063 | 0.05 |
ENST00000541929.1
|
HAL
|
histidine ammonia-lyase |
| chr16_+_2570340 | 0.05 |
ENST00000568263.1
ENST00000293971.6 ENST00000302956.4 ENST00000413459.3 ENST00000566706.1 ENST00000569879.1 |
AMDHD2
|
amidohydrolase domain containing 2 |
| chr14_-_24898731 | 0.05 |
ENST00000267406.6
|
CBLN3
|
cerebellin 3 precursor |
| chr1_+_158323755 | 0.05 |
ENST00000368157.1
ENST00000368156.1 ENST00000368155.3 ENST00000368154.1 ENST00000368160.3 ENST00000368161.3 |
CD1E
|
CD1e molecule |
| chr7_-_99569468 | 0.05 |
ENST00000419575.1
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding |
Network of associatons between targets according to the STRING database.
First level regulatory network of PPARG
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.6 | GO:1904199 | positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.1 | 0.4 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.1 | 0.2 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.2 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.1 | 0.4 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.1 | 0.2 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.2 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.5 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.2 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.1 | GO:0061528 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.0 | 0.1 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.2 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.1 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.0 | 0.2 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.1 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.7 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.1 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.1 | GO:0051801 | cytolysis in other organism involved in symbiotic interaction(GO:0051801) |
| 0.0 | 0.2 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.2 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:0002353 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.0 | 0.1 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.4 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.2 | GO:0018095 | sperm axoneme assembly(GO:0007288) protein polyglutamylation(GO:0018095) |
| 0.0 | 0.0 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.0 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.0 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.0 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.0 | 0.2 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.1 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.0 | 0.0 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.2 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.1 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.0 | GO:0070781 | response to biotin(GO:0070781) |
| 0.0 | 0.2 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0050668 | cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.0 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.3 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.2 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0097342 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.1 | 0.2 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.2 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.1 | 0.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 2.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.2 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.2 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.2 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.0 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.0 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.0 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.0 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.0 | GO:0031177 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.0 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.0 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |