Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
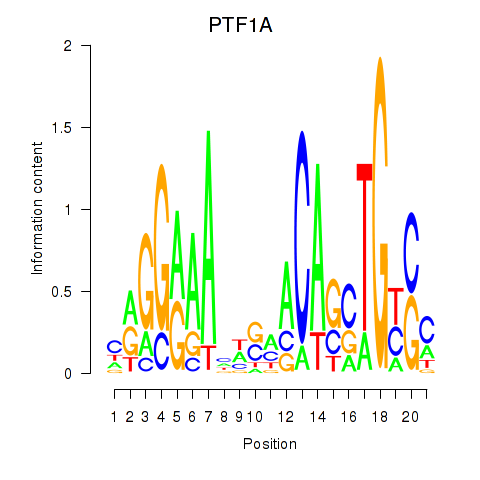
Results for PTF1A
Z-value: 0.77
Transcription factors associated with PTF1A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PTF1A
|
ENSG00000168267.5 | pancreas associated transcription factor 1a |
Activity profile of PTF1A motif
Sorted Z-values of PTF1A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_71238313 | 3.11 |
ENST00000398536.4
|
KRTAP5-7
|
keratin associated protein 5-7 |
| chr4_+_156587853 | 2.19 |
ENST00000506455.1
ENST00000511108.1 |
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr4_+_156588115 | 2.14 |
ENST00000455639.2
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr21_-_46012386 | 2.07 |
ENST00000400368.1
|
KRTAP10-6
|
keratin associated protein 10-6 |
| chr4_+_156587979 | 2.01 |
ENST00000511507.1
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr7_+_150688166 | 1.96 |
ENST00000461406.1
|
NOS3
|
nitric oxide synthase 3 (endothelial cell) |
| chr20_+_42544782 | 1.70 |
ENST00000423191.2
ENST00000372999.1 |
TOX2
|
TOX high mobility group box family member 2 |
| chr4_-_123843597 | 1.66 |
ENST00000510735.1
ENST00000304430.5 |
NUDT6
|
nudix (nucleoside diphosphate linked moiety X)-type motif 6 |
| chr11_+_7597639 | 1.54 |
ENST00000533792.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr11_-_1629693 | 1.37 |
ENST00000399685.1
|
KRTAP5-3
|
keratin associated protein 5-3 |
| chr2_-_241622278 | 1.10 |
ENST00000407834.3
|
AQP12B
|
aquaporin 12B |
| chr1_+_231114795 | 1.01 |
ENST00000310256.2
ENST00000366658.2 ENST00000450711.1 ENST00000435927.1 |
ARV1
|
ARV1 homolog (S. cerevisiae) |
| chr2_+_241631262 | 1.00 |
ENST00000337801.4
ENST00000429564.1 |
AQP12A
|
aquaporin 12A |
| chr4_-_89744457 | 0.98 |
ENST00000395002.2
|
FAM13A
|
family with sequence similarity 13, member A |
| chr12_+_109826524 | 0.96 |
ENST00000431443.2
|
MYO1H
|
myosin IH |
| chr4_+_40198527 | 0.96 |
ENST00000381799.5
|
RHOH
|
ras homolog family member H |
| chr1_-_231114542 | 0.95 |
ENST00000522821.1
ENST00000366661.4 ENST00000366662.4 ENST00000414259.1 ENST00000522399.1 |
TTC13
|
tetratricopeptide repeat domain 13 |
| chr5_-_39425222 | 0.90 |
ENST00000320816.6
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr5_-_39425290 | 0.89 |
ENST00000545653.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr19_-_10491130 | 0.87 |
ENST00000530829.1
ENST00000529370.1 |
TYK2
|
tyrosine kinase 2 |
| chr1_-_33647267 | 0.87 |
ENST00000291416.5
|
TRIM62
|
tripartite motif containing 62 |
| chr17_-_73892992 | 0.85 |
ENST00000540128.1
ENST00000269383.3 |
TRIM65
|
tripartite motif containing 65 |
| chr17_+_67498538 | 0.83 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr5_-_39425068 | 0.83 |
ENST00000515700.1
ENST00000339788.6 |
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr1_-_76076793 | 0.81 |
ENST00000370859.3
|
SLC44A5
|
solute carrier family 44, member 5 |
| chr5_+_78532003 | 0.75 |
ENST00000396137.4
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr4_-_83719983 | 0.75 |
ENST00000319540.4
|
SCD5
|
stearoyl-CoA desaturase 5 |
| chr1_-_205649580 | 0.73 |
ENST00000367145.3
|
SLC45A3
|
solute carrier family 45, member 3 |
| chr11_-_1643368 | 0.70 |
ENST00000399682.1
|
KRTAP5-4
|
keratin associated protein 5-4 |
| chr19_-_46526304 | 0.69 |
ENST00000008938.4
|
PGLYRP1
|
peptidoglycan recognition protein 1 |
| chr4_+_81951957 | 0.69 |
ENST00000282701.2
|
BMP3
|
bone morphogenetic protein 3 |
| chr6_-_2962331 | 0.67 |
ENST00000380524.1
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| chr8_-_99306564 | 0.67 |
ENST00000430223.2
|
NIPAL2
|
NIPA-like domain containing 2 |
| chr18_-_53177984 | 0.66 |
ENST00000543082.1
|
TCF4
|
transcription factor 4 |
| chr1_-_9129598 | 0.66 |
ENST00000535586.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr13_-_79979952 | 0.63 |
ENST00000438724.1
|
RBM26
|
RNA binding motif protein 26 |
| chr13_-_79979919 | 0.59 |
ENST00000267229.7
|
RBM26
|
RNA binding motif protein 26 |
| chr17_-_34313685 | 0.59 |
ENST00000435911.2
ENST00000586216.1 ENST00000394509.4 |
CCL14
|
chemokine (C-C motif) ligand 14 |
| chr2_+_202122703 | 0.58 |
ENST00000447616.1
ENST00000358485.4 |
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr5_-_162887071 | 0.58 |
ENST00000302764.4
|
NUDCD2
|
NudC domain containing 2 |
| chr5_-_162887054 | 0.57 |
ENST00000517501.1
|
NUDCD2
|
NudC domain containing 2 |
| chr1_+_178694300 | 0.56 |
ENST00000367635.3
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr3_-_169587621 | 0.52 |
ENST00000523069.1
ENST00000316428.5 ENST00000264676.5 |
LRRC31
|
leucine rich repeat containing 31 |
| chr11_+_64107663 | 0.51 |
ENST00000356786.5
|
CCDC88B
|
coiled-coil domain containing 88B |
| chr1_-_78149041 | 0.51 |
ENST00000414381.1
ENST00000370798.1 |
ZZZ3
|
zinc finger, ZZ-type containing 3 |
| chr1_-_9129895 | 0.51 |
ENST00000473209.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr8_-_99306611 | 0.50 |
ENST00000341166.3
|
NIPAL2
|
NIPA-like domain containing 2 |
| chrX_-_107019181 | 0.50 |
ENST00000315660.4
ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr1_+_26856236 | 0.48 |
ENST00000374168.2
ENST00000374166.4 |
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr20_+_18125727 | 0.47 |
ENST00000489634.2
|
CSRP2BP
|
CSRP2 binding protein |
| chr3_+_49027308 | 0.47 |
ENST00000383729.4
ENST00000343546.4 |
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr12_+_50451331 | 0.45 |
ENST00000228468.4
|
ASIC1
|
acid-sensing (proton-gated) ion channel 1 |
| chr5_-_75013193 | 0.44 |
ENST00000514838.2
ENST00000506164.1 ENST00000502826.1 ENST00000503835.1 ENST00000428202.2 ENST00000380475.2 |
POC5
|
POC5 centriolar protein |
| chr1_-_114447620 | 0.44 |
ENST00000369569.1
ENST00000432415.1 ENST00000369571.2 |
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit |
| chr1_-_45956822 | 0.43 |
ENST00000372086.3
ENST00000341771.6 |
TESK2
|
testis-specific kinase 2 |
| chr15_+_41099788 | 0.43 |
ENST00000299173.10
ENST00000566407.1 |
ZFYVE19
|
zinc finger, FYVE domain containing 19 |
| chr8_-_38126635 | 0.42 |
ENST00000529359.1
|
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B |
| chr9_+_138235095 | 0.42 |
ENST00000320778.2
|
C9orf62
|
chromosome 9 open reading frame 62 |
| chr19_+_6464502 | 0.42 |
ENST00000308243.7
|
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr6_-_159466042 | 0.41 |
ENST00000338313.5
|
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chr1_-_76076759 | 0.41 |
ENST00000370855.5
|
SLC44A5
|
solute carrier family 44, member 5 |
| chr20_+_54823788 | 0.40 |
ENST00000243911.2
|
MC3R
|
melanocortin 3 receptor |
| chr12_-_90102594 | 0.40 |
ENST00000428670.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr9_-_116061145 | 0.39 |
ENST00000416588.2
|
RNF183
|
ring finger protein 183 |
| chr1_-_114447683 | 0.38 |
ENST00000256658.4
ENST00000369564.1 |
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit |
| chr7_-_142207004 | 0.38 |
ENST00000426318.2
|
TRBV10-2
|
T cell receptor beta variable 10-2 |
| chr4_-_83719884 | 0.36 |
ENST00000282709.4
ENST00000273908.4 |
SCD5
|
stearoyl-CoA desaturase 5 |
| chr19_-_39694894 | 0.35 |
ENST00000318438.6
|
SYCN
|
syncollin |
| chr19_+_54495542 | 0.34 |
ENST00000252729.2
ENST00000352529.1 |
CACNG6
|
calcium channel, voltage-dependent, gamma subunit 6 |
| chr2_-_89278535 | 0.34 |
ENST00000390247.2
|
IGKV3-7
|
immunoglobulin kappa variable 3-7 (non-functional) |
| chr4_-_14889791 | 0.34 |
ENST00000509654.1
ENST00000515031.1 ENST00000505089.2 |
LINC00504
|
long intergenic non-protein coding RNA 504 |
| chr12_-_48398104 | 0.33 |
ENST00000337299.6
ENST00000380518.3 |
COL2A1
|
collagen, type II, alpha 1 |
| chr6_+_57182400 | 0.33 |
ENST00000607273.1
|
PRIM2
|
primase, DNA, polypeptide 2 (58kDa) |
| chr13_-_31736027 | 0.32 |
ENST00000380406.5
ENST00000320027.5 ENST00000380405.4 |
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr10_+_124739964 | 0.32 |
ENST00000406217.2
|
PSTK
|
phosphoseryl-tRNA kinase |
| chr12_+_75784850 | 0.31 |
ENST00000550916.1
ENST00000435775.1 ENST00000378689.2 ENST00000378692.3 ENST00000320460.4 ENST00000547164.1 |
GLIPR1L2
|
GLI pathogenesis-related 1 like 2 |
| chr1_-_114447412 | 0.29 |
ENST00000369567.1
ENST00000369566.3 |
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit |
| chr12_+_112451120 | 0.29 |
ENST00000261735.3
ENST00000455836.1 |
ERP29
|
endoplasmic reticulum protein 29 |
| chr1_+_150480576 | 0.29 |
ENST00000346569.6
|
ECM1
|
extracellular matrix protein 1 |
| chr8_+_11351494 | 0.29 |
ENST00000259089.4
|
BLK
|
B lymphoid tyrosine kinase |
| chr7_+_141811539 | 0.28 |
ENST00000550469.2
ENST00000477922.3 |
RP11-1220K2.2
|
Putative inactive maltase-glucoamylase-like protein LOC93432 |
| chr20_-_44259893 | 0.28 |
ENST00000326000.1
|
WFDC9
|
WAP four-disulfide core domain 9 |
| chr9_+_125281420 | 0.27 |
ENST00000340750.1
|
OR1J4
|
olfactory receptor, family 1, subfamily J, member 4 |
| chr10_+_124739911 | 0.26 |
ENST00000405485.1
|
PSTK
|
phosphoseryl-tRNA kinase |
| chr17_-_3461092 | 0.26 |
ENST00000301365.4
ENST00000572519.1 |
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3 |
| chr6_-_41715128 | 0.26 |
ENST00000356667.4
ENST00000373025.3 ENST00000425343.2 |
PGC
|
progastricsin (pepsinogen C) |
| chrX_-_48216101 | 0.25 |
ENST00000298396.2
ENST00000376893.3 |
SSX3
|
synovial sarcoma, X breakpoint 3 |
| chr1_+_150480551 | 0.25 |
ENST00000369049.4
ENST00000369047.4 |
ECM1
|
extracellular matrix protein 1 |
| chr11_-_111794446 | 0.24 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr6_-_56258892 | 0.24 |
ENST00000370819.1
|
COL21A1
|
collagen, type XXI, alpha 1 |
| chr20_+_44637526 | 0.24 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr1_+_12185949 | 0.23 |
ENST00000413146.2
|
TNFRSF8
|
tumor necrosis factor receptor superfamily, member 8 |
| chr17_+_27895609 | 0.23 |
ENST00000581411.2
ENST00000301057.7 |
TP53I13
|
tumor protein p53 inducible protein 13 |
| chrX_-_44402231 | 0.23 |
ENST00000378045.4
|
FUNDC1
|
FUN14 domain containing 1 |
| chr1_+_151682909 | 0.22 |
ENST00000326413.3
ENST00000442233.2 |
RIIAD1
AL589765.1
|
regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1 Uncharacterized protein; cDNA FLJ36032 fis, clone TESTI2017069 |
| chr4_-_89744314 | 0.22 |
ENST00000508369.1
|
FAM13A
|
family with sequence similarity 13, member A |
| chr13_+_113633620 | 0.22 |
ENST00000421756.1
ENST00000375601.3 |
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr7_-_152373216 | 0.21 |
ENST00000359321.1
|
XRCC2
|
X-ray repair complementing defective repair in Chinese hamster cells 2 |
| chr3_-_88108192 | 0.21 |
ENST00000309534.6
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr22_-_17640110 | 0.20 |
ENST00000399852.3
ENST00000336737.4 |
CECR5
|
cat eye syndrome chromosome region, candidate 5 |
| chr2_-_27435125 | 0.20 |
ENST00000414408.1
ENST00000310574.3 |
SLC5A6
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chrX_+_154299690 | 0.20 |
ENST00000340647.4
ENST00000330045.7 |
BRCC3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chr2_+_173600514 | 0.20 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr12_+_53689309 | 0.19 |
ENST00000351500.3
ENST00000550846.1 ENST00000334478.4 ENST00000549759.1 |
PFDN5
|
prefoldin subunit 5 |
| chr14_-_50154921 | 0.19 |
ENST00000553805.2
ENST00000554396.1 ENST00000216367.5 ENST00000539565.2 |
POLE2
|
polymerase (DNA directed), epsilon 2, accessory subunit |
| chr14_+_77292715 | 0.19 |
ENST00000393774.3
ENST00000555189.1 ENST00000450042.2 |
C14orf166B
|
chromosome 14 open reading frame 166B |
| chr2_+_173600565 | 0.19 |
ENST00000397081.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr1_+_32084794 | 0.18 |
ENST00000373705.1
|
HCRTR1
|
hypocretin (orexin) receptor 1 |
| chr10_+_76871454 | 0.18 |
ENST00000372687.4
|
SAMD8
|
sterile alpha motif domain containing 8 |
| chr14_-_77737543 | 0.18 |
ENST00000298352.4
|
NGB
|
neuroglobin |
| chr8_+_11351876 | 0.18 |
ENST00000529894.1
|
BLK
|
B lymphoid tyrosine kinase |
| chr4_-_89744365 | 0.17 |
ENST00000513837.1
ENST00000503556.1 |
FAM13A
|
family with sequence similarity 13, member A |
| chr3_+_40518599 | 0.17 |
ENST00000314686.5
ENST00000447116.2 ENST00000429348.2 ENST00000456778.1 |
ZNF619
|
zinc finger protein 619 |
| chr2_+_120187465 | 0.17 |
ENST00000409826.1
ENST00000417645.1 |
TMEM37
|
transmembrane protein 37 |
| chrX_-_13956497 | 0.17 |
ENST00000398361.3
|
GPM6B
|
glycoprotein M6B |
| chr16_+_71392616 | 0.16 |
ENST00000349553.5
ENST00000302628.4 ENST00000562305.1 |
CALB2
|
calbindin 2 |
| chr1_+_100503643 | 0.16 |
ENST00000370152.3
|
HIAT1
|
hippocampus abundant transcript 1 |
| chr12_-_56223394 | 0.15 |
ENST00000317269.3
|
DNAJC14
|
DnaJ (Hsp40) homolog, subfamily C, member 14 |
| chr13_+_113812956 | 0.14 |
ENST00000375547.2
ENST00000342783.4 |
PROZ
|
protein Z, vitamin K-dependent plasma glycoprotein |
| chr2_+_27435179 | 0.14 |
ENST00000606999.1
ENST00000405489.3 |
ATRAID
|
all-trans retinoic acid-induced differentiation factor |
| chr20_+_29956369 | 0.13 |
ENST00000253381.2
|
DEFB118
|
defensin, beta 118 |
| chr17_+_53342311 | 0.13 |
ENST00000226067.5
|
HLF
|
hepatic leukemia factor |
| chr17_+_12692774 | 0.13 |
ENST00000379672.5
ENST00000340825.3 |
ARHGAP44
|
Rho GTPase activating protein 44 |
| chrX_-_13956737 | 0.13 |
ENST00000454189.2
|
GPM6B
|
glycoprotein M6B |
| chrX_+_154299753 | 0.13 |
ENST00000369459.2
ENST00000369462.1 ENST00000411985.1 ENST00000399042.1 |
BRCC3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chr22_-_39190116 | 0.13 |
ENST00000406622.1
ENST00000216068.4 ENST00000406199.3 |
SUN2
DNAL4
|
Sad1 and UNC84 domain containing 2 dynein, axonemal, light chain 4 |
| chr16_+_89696692 | 0.13 |
ENST00000261615.4
|
DPEP1
|
dipeptidase 1 (renal) |
| chr6_-_24877490 | 0.13 |
ENST00000540914.1
ENST00000378023.4 |
FAM65B
|
family with sequence similarity 65, member B |
| chr14_-_35099377 | 0.13 |
ENST00000362031.4
|
SNX6
|
sorting nexin 6 |
| chr15_+_68924327 | 0.12 |
ENST00000543950.1
|
CORO2B
|
coronin, actin binding protein, 2B |
| chr13_+_107028897 | 0.11 |
ENST00000439790.1
ENST00000435024.1 |
LINC00460
|
long intergenic non-protein coding RNA 460 |
| chr11_+_64058758 | 0.10 |
ENST00000538767.1
|
KCNK4
|
potassium channel, subfamily K, member 4 |
| chr16_-_15950868 | 0.10 |
ENST00000396324.3
ENST00000452625.2 ENST00000576790.2 ENST00000300036.5 |
MYH11
|
myosin, heavy chain 11, smooth muscle |
| chr16_+_2802316 | 0.10 |
ENST00000301740.8
|
SRRM2
|
serine/arginine repetitive matrix 2 |
| chr18_+_5238055 | 0.09 |
ENST00000582363.1
ENST00000582008.1 ENST00000580082.1 |
LINC00667
|
long intergenic non-protein coding RNA 667 |
| chr8_-_38126675 | 0.09 |
ENST00000531823.1
ENST00000534339.1 ENST00000524616.1 ENST00000422581.2 ENST00000424479.2 ENST00000419686.2 |
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B |
| chr1_-_45956800 | 0.09 |
ENST00000538496.1
|
TESK2
|
testis-specific kinase 2 |
| chr16_-_2004683 | 0.09 |
ENST00000268661.7
|
RPL3L
|
ribosomal protein L3-like |
| chr10_+_120116527 | 0.08 |
ENST00000445161.1
|
LINC00867
|
long intergenic non-protein coding RNA 867 |
| chr19_+_11670245 | 0.08 |
ENST00000585493.1
|
ZNF627
|
zinc finger protein 627 |
| chr12_-_50677255 | 0.08 |
ENST00000551691.1
ENST00000394943.3 ENST00000341247.4 |
LIMA1
|
LIM domain and actin binding 1 |
| chr13_-_31736132 | 0.08 |
ENST00000429785.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr3_-_12883026 | 0.08 |
ENST00000396953.2
ENST00000457131.1 ENST00000435983.1 ENST00000273223.6 ENST00000396957.1 ENST00000429711.2 |
RPL32
|
ribosomal protein L32 |
| chr2_-_136288113 | 0.08 |
ENST00000401392.1
|
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr2_-_89417335 | 0.07 |
ENST00000490686.1
|
IGKV1-17
|
immunoglobulin kappa variable 1-17 |
| chr6_-_159466136 | 0.07 |
ENST00000367066.3
ENST00000326965.6 |
TAGAP
|
T-cell activation RhoGTPase activating protein |
| chr19_+_6464243 | 0.07 |
ENST00000600229.1
ENST00000356762.3 |
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr10_+_43572475 | 0.07 |
ENST00000355710.3
ENST00000498820.1 ENST00000340058.5 |
RET
|
ret proto-oncogene |
| chr11_-_130298888 | 0.06 |
ENST00000257359.6
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8 |
| chrX_+_155110956 | 0.06 |
ENST00000286448.6
ENST00000262640.6 ENST00000460621.1 |
VAMP7
|
vesicle-associated membrane protein 7 |
| chr9_-_104145795 | 0.06 |
ENST00000259407.2
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) |
| chr14_-_35099315 | 0.06 |
ENST00000396526.3
ENST00000396534.3 ENST00000355110.5 ENST00000557265.1 |
SNX6
|
sorting nexin 6 |
| chr3_-_193272874 | 0.06 |
ENST00000342695.4
|
ATP13A4
|
ATPase type 13A4 |
| chr1_+_40204538 | 0.06 |
ENST00000324379.5
ENST00000356511.2 ENST00000497370.1 ENST00000470213.1 ENST00000372835.5 ENST00000372830.1 |
PPIE
|
peptidylprolyl isomerase E (cyclophilin E) |
| chr12_+_1929421 | 0.06 |
ENST00000543818.1
|
LRTM2
|
leucine-rich repeats and transmembrane domains 2 |
| chr22_+_24999114 | 0.05 |
ENST00000412658.1
ENST00000445029.1 ENST00000419133.1 ENST00000400382.1 ENST00000438643.2 ENST00000452551.1 ENST00000400383.1 ENST00000412898.1 ENST00000400380.1 ENST00000455483.1 ENST00000430289.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr4_+_20702059 | 0.05 |
ENST00000444671.2
ENST00000510700.1 ENST00000506745.1 ENST00000514663.1 ENST00000509469.1 ENST00000515339.1 ENST00000513861.1 ENST00000502374.1 ENST00000538990.1 ENST00000511160.1 ENST00000504630.1 ENST00000513590.1 ENST00000514292.1 ENST00000502938.1 ENST00000509625.1 ENST00000505160.1 ENST00000507634.1 ENST00000513459.1 ENST00000511089.1 |
PACRGL
|
PARK2 co-regulated-like |
| chr7_+_142458507 | 0.05 |
ENST00000492062.1
|
PRSS1
|
protease, serine, 1 (trypsin 1) |
| chr22_+_23101182 | 0.05 |
ENST00000390312.2
|
IGLV2-14
|
immunoglobulin lambda variable 2-14 |
| chr6_-_46620522 | 0.05 |
ENST00000275016.2
|
CYP39A1
|
cytochrome P450, family 39, subfamily A, polypeptide 1 |
| chr12_-_120884175 | 0.05 |
ENST00000546954.1
|
TRIAP1
|
TP53 regulated inhibitor of apoptosis 1 |
| chr9_-_138853156 | 0.05 |
ENST00000371756.3
|
UBAC1
|
UBA domain containing 1 |
| chr7_+_30323923 | 0.04 |
ENST00000323037.4
|
ZNRF2
|
zinc and ring finger 2 |
| chr9_-_19033185 | 0.04 |
ENST00000380534.4
|
FAM154A
|
family with sequence similarity 154, member A |
| chr1_+_228337553 | 0.04 |
ENST00000366714.2
|
GJC2
|
gap junction protein, gamma 2, 47kDa |
| chr9_-_19033237 | 0.04 |
ENST00000380530.1
ENST00000542071.1 |
FAM154A
|
family with sequence similarity 154, member A |
| chr11_+_56949221 | 0.04 |
ENST00000497933.1
|
LRRC55
|
leucine rich repeat containing 55 |
| chr8_-_135725205 | 0.03 |
ENST00000523399.1
ENST00000377838.3 |
ZFAT
|
zinc finger and AT hook domain containing |
| chr17_+_40996590 | 0.03 |
ENST00000253799.3
ENST00000452774.2 |
AOC2
|
amine oxidase, copper containing 2 (retina-specific) |
| chr20_-_48732472 | 0.03 |
ENST00000340309.3
ENST00000415862.2 ENST00000371677.3 ENST00000420027.2 |
UBE2V1
|
ubiquitin-conjugating enzyme E2 variant 1 |
| chr1_-_158300747 | 0.03 |
ENST00000451207.1
|
CD1B
|
CD1b molecule |
| chr6_-_24489842 | 0.03 |
ENST00000230036.1
|
GPLD1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr11_+_8704748 | 0.03 |
ENST00000526562.1
ENST00000525981.1 |
RPL27A
|
ribosomal protein L27a |
| chr18_+_33877654 | 0.02 |
ENST00000257209.4
ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3
|
formin homology 2 domain containing 3 |
| chr1_+_32084641 | 0.02 |
ENST00000373706.5
|
HCRTR1
|
hypocretin (orexin) receptor 1 |
| chr1_-_9129735 | 0.02 |
ENST00000377424.4
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr19_+_42300548 | 0.02 |
ENST00000344550.4
|
CEACAM3
|
carcinoembryonic antigen-related cell adhesion molecule 3 |
| chr2_+_183943464 | 0.02 |
ENST00000354221.4
|
DUSP19
|
dual specificity phosphatase 19 |
| chr17_+_31318886 | 0.01 |
ENST00000269053.3
ENST00000394638.1 |
SPACA3
|
sperm acrosome associated 3 |
| chr12_+_1929644 | 0.01 |
ENST00000299194.1
|
LRTM2
|
leucine-rich repeats and transmembrane domains 2 |
| chr11_-_63933504 | 0.01 |
ENST00000255681.6
|
MACROD1
|
MACRO domain containing 1 |
| chr8_-_145641864 | 0.01 |
ENST00000276833.5
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr19_+_19779686 | 0.00 |
ENST00000415784.2
|
ZNF101
|
zinc finger protein 101 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PTF1A
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.3 | GO:0052551 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.7 | 2.6 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.7 | 2.0 | GO:0014806 | negative regulation of muscle hyperplasia(GO:0014740) smooth muscle hyperplasia(GO:0014806) |
| 0.4 | 1.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.3 | 0.8 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.2 | 0.7 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.2 | 0.7 | GO:0002325 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.2 | 1.0 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.2 | 0.8 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.6 | GO:0052056 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.1 | 1.0 | GO:0098704 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.1 | 0.6 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.6 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.1 | 1.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.2 | GO:0032679 | TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.1 | 0.3 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.1 | 1.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 0.4 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.1 | 0.5 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.5 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 0.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.5 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.3 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 0.3 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.1 | 0.4 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 0.4 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.3 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.1 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.0 | 0.2 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 6.8 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.2 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 0.3 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:0071469 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.3 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.2 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.4 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.9 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0007497 | posterior midgut development(GO:0007497) |
| 0.0 | 0.5 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 1.0 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 1.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.7 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.6 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.6 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 6.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 1.0 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.6 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.1 | 6.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.3 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.6 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 1.1 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.4 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 1.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.2 | 0.7 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 6.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 1.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.2 | 0.7 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.1 | 1.0 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.1 | 1.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 2.6 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.4 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.6 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 0.3 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 0.5 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.2 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 0.5 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 1.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.5 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.7 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.2 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.1 | 0.7 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 1.0 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 1.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.0 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.3 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 2.0 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 2.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.5 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 6.2 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 1.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.6 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.8 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.3 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 2.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |