Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
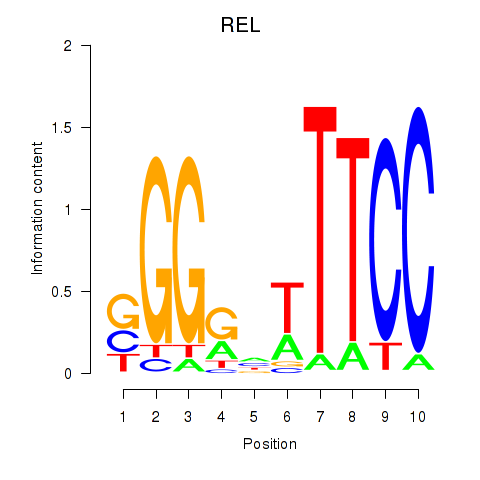
Results for REL
Z-value: 2.66
Transcription factors associated with REL
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
REL
|
ENSG00000162924.9 | REL proto-oncogene, NF-kB subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| REL | hg19_v2_chr2_+_61108650_61108687 | -0.16 | 4.6e-01 | Click! |
Activity profile of REL motif
Sorted Z-values of REL motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_29527702 | 28.30 |
ENST00000377050.4
|
UBD
|
ubiquitin D |
| chr6_-_31550192 | 21.69 |
ENST00000429299.2
ENST00000446745.2 |
LTB
|
lymphotoxin beta (TNF superfamily, member 3) |
| chr4_+_74702214 | 19.95 |
ENST00000226317.5
ENST00000515050.1 |
CXCL6
|
chemokine (C-X-C motif) ligand 6 |
| chr6_+_138188551 | 18.48 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr4_-_76957214 | 17.31 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr19_+_4229495 | 15.93 |
ENST00000221847.5
|
EBI3
|
Epstein-Barr virus induced 3 |
| chr5_-_150460914 | 15.22 |
ENST00000389378.2
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr11_+_102188224 | 15.07 |
ENST00000263464.3
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr10_+_104155450 | 14.80 |
ENST00000471698.1
ENST00000189444.6 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr2_+_228678550 | 14.53 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr11_+_19798964 | 14.44 |
ENST00000527559.2
|
NAV2
|
neuron navigator 2 |
| chr11_+_102188272 | 14.33 |
ENST00000532808.1
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr14_+_103589789 | 13.77 |
ENST00000558056.1
ENST00000560869.1 |
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr5_-_150466692 | 13.38 |
ENST00000315050.7
ENST00000523338.1 ENST00000522100.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr11_+_19799327 | 13.36 |
ENST00000540292.1
|
NAV2
|
neuron navigator 2 |
| chr4_-_74864386 | 13.19 |
ENST00000296027.4
|
CXCL5
|
chemokine (C-X-C motif) ligand 5 |
| chr1_-_169703203 | 13.08 |
ENST00000333360.7
ENST00000367781.4 ENST00000367782.4 ENST00000367780.4 ENST00000367779.4 |
SELE
|
selectin E |
| chr1_-_8000872 | 12.43 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr15_+_67430339 | 11.17 |
ENST00000439724.3
|
SMAD3
|
SMAD family member 3 |
| chr1_+_110453109 | 10.68 |
ENST00000525659.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr6_-_44233361 | 10.00 |
ENST00000275015.5
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| chr12_+_102271129 | 9.36 |
ENST00000258534.8
|
DRAM1
|
DNA-damage regulated autophagy modulator 1 |
| chr1_+_110453203 | 9.11 |
ENST00000357302.4
ENST00000344188.5 ENST00000329608.6 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr20_+_44746885 | 8.96 |
ENST00000372285.3
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chr20_+_44746939 | 8.68 |
ENST00000372276.3
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chr10_+_104154229 | 8.68 |
ENST00000428099.1
ENST00000369966.3 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr2_-_204400113 | 8.38 |
ENST00000319170.5
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr12_-_57504069 | 8.36 |
ENST00000543873.2
ENST00000554663.1 ENST00000557635.1 |
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr6_-_32821599 | 8.29 |
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr8_+_123793633 | 8.23 |
ENST00000314393.4
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr6_+_32821924 | 8.03 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr9_-_136344197 | 7.78 |
ENST00000414172.1
ENST00000371897.4 |
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr11_-_72492903 | 7.45 |
ENST00000537947.1
|
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr1_+_110453462 | 7.33 |
ENST00000488198.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr11_-_72492878 | 7.23 |
ENST00000535054.1
ENST00000545082.1 |
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr4_+_74735102 | 7.18 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr8_-_119964434 | 6.78 |
ENST00000297350.4
|
TNFRSF11B
|
tumor necrosis factor receptor superfamily, member 11b |
| chr9_+_72658490 | 6.57 |
ENST00000377182.4
|
MAMDC2
|
MAM domain containing 2 |
| chr11_-_128392085 | 6.48 |
ENST00000526145.2
ENST00000531611.1 ENST00000319397.6 ENST00000345075.4 ENST00000535549.1 |
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr19_-_47734448 | 6.37 |
ENST00000439096.2
|
BBC3
|
BCL2 binding component 3 |
| chr16_+_50730910 | 6.11 |
ENST00000300589.2
|
NOD2
|
nucleotide-binding oligomerization domain containing 2 |
| chr22_-_19512893 | 6.11 |
ENST00000403084.1
ENST00000413119.2 |
CLDN5
|
claudin 5 |
| chr7_-_47579188 | 6.08 |
ENST00000398879.1
ENST00000355730.3 ENST00000442536.2 ENST00000458317.2 |
TNS3
|
tensin 3 |
| chr12_-_77272765 | 6.07 |
ENST00000547435.1
ENST00000552330.1 ENST00000546966.1 ENST00000311083.5 |
CSRP2
|
cysteine and glycine-rich protein 2 |
| chr17_+_40440481 | 5.99 |
ENST00000590726.2
ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A
|
signal transducer and activator of transcription 5A |
| chr19_-_4338783 | 5.87 |
ENST00000601482.1
ENST00000600324.1 |
STAP2
|
signal transducing adaptor family member 2 |
| chr21_+_34775772 | 5.85 |
ENST00000405436.1
|
IFNGR2
|
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr19_-_4338838 | 5.84 |
ENST00000594605.1
|
STAP2
|
signal transducing adaptor family member 2 |
| chr21_+_34775698 | 5.61 |
ENST00000381995.1
|
IFNGR2
|
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr21_+_34775181 | 5.46 |
ENST00000290219.6
|
IFNGR2
|
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr17_+_26662730 | 5.17 |
ENST00000226225.2
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr1_+_165796753 | 5.14 |
ENST00000367879.4
|
UCK2
|
uridine-cytidine kinase 2 |
| chr2_-_163175133 | 5.13 |
ENST00000421365.2
ENST00000263642.2 |
IFIH1
|
interferon induced with helicase C domain 1 |
| chr2_+_162016827 | 5.10 |
ENST00000429217.1
ENST00000406287.1 ENST00000402568.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr10_+_90750378 | 4.96 |
ENST00000355740.2
ENST00000352159.4 |
FAS
|
Fas cell surface death receptor |
| chr1_+_144339738 | 4.92 |
ENST00000538264.1
|
AL592284.1
|
Protein LOC642441 |
| chr14_-_24616426 | 4.88 |
ENST00000216802.5
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr15_-_51058005 | 4.88 |
ENST00000261854.5
|
SPPL2A
|
signal peptide peptidase like 2A |
| chr20_-_43977055 | 4.83 |
ENST00000372733.3
ENST00000537976.1 |
SDC4
|
syndecan 4 |
| chr10_+_90750493 | 4.51 |
ENST00000357339.2
ENST00000355279.2 |
FAS
|
Fas cell surface death receptor |
| chr5_+_14143728 | 4.44 |
ENST00000344204.4
ENST00000537187.1 |
TRIO
|
trio Rho guanine nucleotide exchange factor |
| chr1_-_209824643 | 4.43 |
ENST00000391911.1
ENST00000415782.1 |
LAMB3
|
laminin, beta 3 |
| chr14_+_103243813 | 4.38 |
ENST00000560371.1
ENST00000347662.4 ENST00000392745.2 ENST00000539721.1 ENST00000560463.1 |
TRAF3
|
TNF receptor-associated factor 3 |
| chr12_-_9913489 | 4.38 |
ENST00000228434.3
ENST00000536709.1 |
CD69
|
CD69 molecule |
| chr2_+_102314161 | 4.35 |
ENST00000425019.1
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr2_+_162016916 | 4.35 |
ENST00000405852.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr10_-_96122682 | 4.34 |
ENST00000371361.3
|
NOC3L
|
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr16_+_31044812 | 4.00 |
ENST00000313843.3
|
STX4
|
syntaxin 4 |
| chr7_-_98741714 | 3.95 |
ENST00000361125.1
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1 |
| chr7_-_98741642 | 3.93 |
ENST00000361368.2
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1 |
| chr4_-_185395672 | 3.88 |
ENST00000393593.3
|
IRF2
|
interferon regulatory factor 2 |
| chr1_-_65432171 | 3.85 |
ENST00000342505.4
|
JAK1
|
Janus kinase 1 |
| chr19_-_17958832 | 3.81 |
ENST00000458235.1
|
JAK3
|
Janus kinase 3 |
| chr2_-_128145498 | 3.79 |
ENST00000409179.2
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr11_-_75062829 | 3.73 |
ENST00000393505.4
|
ARRB1
|
arrestin, beta 1 |
| chr10_+_13142075 | 3.62 |
ENST00000378757.2
ENST00000430081.1 ENST00000378752.3 ENST00000378748.3 |
OPTN
|
optineurin |
| chr5_+_133984462 | 3.56 |
ENST00000398844.2
ENST00000322887.4 |
SEC24A
|
SEC24 family member A |
| chr8_-_105601134 | 3.52 |
ENST00000276654.5
ENST00000424843.2 |
LRP12
|
low density lipoprotein receptor-related protein 12 |
| chr10_+_13142225 | 3.51 |
ENST00000378747.3
|
OPTN
|
optineurin |
| chr1_-_204380919 | 3.47 |
ENST00000367188.4
|
PPP1R15B
|
protein phosphatase 1, regulatory subunit 15B |
| chr19_+_39390320 | 3.36 |
ENST00000576510.1
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr10_+_13141585 | 3.35 |
ENST00000378764.2
|
OPTN
|
optineurin |
| chr2_+_68694678 | 3.32 |
ENST00000303795.4
|
APLF
|
aprataxin and PNKP like factor |
| chr13_+_97928395 | 3.24 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr11_-_75062730 | 3.23 |
ENST00000420843.2
ENST00000360025.3 |
ARRB1
|
arrestin, beta 1 |
| chr10_-_100027943 | 3.06 |
ENST00000260702.3
|
LOXL4
|
lysyl oxidase-like 4 |
| chr15_+_71145578 | 3.03 |
ENST00000544974.2
ENST00000558546.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr10_-_97050777 | 3.02 |
ENST00000329399.6
|
PDLIM1
|
PDZ and LIM domain 1 |
| chr16_+_27325202 | 2.96 |
ENST00000395762.2
ENST00000562142.1 ENST00000561742.1 ENST00000543915.2 ENST00000449195.1 ENST00000380922.3 ENST00000563002.1 |
IL4R
|
interleukin 4 receptor |
| chr16_-_75498553 | 2.86 |
ENST00000569276.1
ENST00000357613.4 ENST00000561878.1 ENST00000566980.1 ENST00000567194.1 |
TMEM170A
RP11-77K12.1
|
transmembrane protein 170A Uncharacterized protein |
| chr1_-_159894319 | 2.84 |
ENST00000320307.4
|
TAGLN2
|
transgelin 2 |
| chr1_-_27816556 | 2.81 |
ENST00000536657.1
|
WASF2
|
WAS protein family, member 2 |
| chr6_+_151042224 | 2.80 |
ENST00000358517.2
|
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr9_-_115095851 | 2.64 |
ENST00000343327.2
|
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr6_+_135502408 | 2.64 |
ENST00000341911.5
ENST00000442647.2 ENST00000316528.8 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr8_-_23712312 | 2.59 |
ENST00000290271.2
|
STC1
|
stanniocalcin 1 |
| chr19_+_39390587 | 2.56 |
ENST00000572515.1
ENST00000392079.3 ENST00000575359.1 ENST00000313582.5 |
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr9_+_132934835 | 2.53 |
ENST00000372398.3
|
NCS1
|
neuronal calcium sensor 1 |
| chr4_-_74904398 | 2.52 |
ENST00000296026.4
|
CXCL3
|
chemokine (C-X-C motif) ligand 3 |
| chr19_+_41725140 | 2.50 |
ENST00000359092.3
|
AXL
|
AXL receptor tyrosine kinase |
| chr7_-_19184929 | 2.49 |
ENST00000275461.3
|
FERD3L
|
Fer3-like bHLH transcription factor |
| chr9_-_115095123 | 2.47 |
ENST00000458258.1
|
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr3_+_9691117 | 2.45 |
ENST00000353332.5
ENST00000420925.1 ENST00000296003.4 ENST00000351233.5 |
MTMR14
|
myotubularin related protein 14 |
| chr15_+_52311398 | 2.43 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr12_+_51632600 | 2.42 |
ENST00000549555.1
ENST00000439799.2 ENST00000425012.2 |
DAZAP2
|
DAZ associated protein 2 |
| chr1_+_16767195 | 2.42 |
ENST00000504551.2
ENST00000457722.2 ENST00000406746.1 ENST00000443980.2 |
NECAP2
|
NECAP endocytosis associated 2 |
| chr15_+_44719970 | 2.39 |
ENST00000558966.1
|
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr9_-_115095883 | 2.34 |
ENST00000450374.1
ENST00000374255.2 ENST00000334318.6 ENST00000374257.1 |
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr17_-_7590745 | 2.34 |
ENST00000514944.1
ENST00000503591.1 ENST00000455263.2 ENST00000420246.2 ENST00000445888.2 ENST00000509690.1 ENST00000604348.1 ENST00000269305.4 |
TP53
|
tumor protein p53 |
| chr19_+_496454 | 2.31 |
ENST00000346144.4
ENST00000215637.3 ENST00000382683.4 |
MADCAM1
|
mucosal vascular addressin cell adhesion molecule 1 |
| chr4_-_146101304 | 2.24 |
ENST00000447906.2
|
OTUD4
|
OTU domain containing 4 |
| chr8_-_101321584 | 2.23 |
ENST00000523167.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr1_-_205912577 | 2.22 |
ENST00000367135.3
ENST00000367134.2 |
SLC26A9
|
solute carrier family 26 (anion exchanger), member 9 |
| chr12_-_49504655 | 2.13 |
ENST00000551782.1
ENST00000267102.8 |
LMBR1L
|
limb development membrane protein 1-like |
| chr9_-_117880477 | 2.08 |
ENST00000534839.1
ENST00000340094.3 ENST00000535648.1 ENST00000346706.3 ENST00000345230.3 ENST00000350763.4 |
TNC
|
tenascin C |
| chr1_+_6845384 | 2.08 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr2_+_114647504 | 2.05 |
ENST00000263238.2
|
ACTR3
|
ARP3 actin-related protein 3 homolog (yeast) |
| chr6_+_29910301 | 2.04 |
ENST00000376809.5
ENST00000376802.2 |
HLA-A
|
major histocompatibility complex, class I, A |
| chr22_-_38380543 | 2.04 |
ENST00000396884.2
|
SOX10
|
SRY (sex determining region Y)-box 10 |
| chr12_+_56211703 | 2.02 |
ENST00000243045.5
ENST00000552672.1 ENST00000550836.1 |
ORMDL2
|
ORM1-like 2 (S. cerevisiae) |
| chrX_-_48755030 | 1.99 |
ENST00000490755.2
ENST00000465150.2 ENST00000495490.2 |
TIMM17B
|
translocase of inner mitochondrial membrane 17 homolog B (yeast) |
| chr2_+_90153696 | 1.96 |
ENST00000417279.2
|
IGKV3D-15
|
immunoglobulin kappa variable 3D-15 (gene/pseudogene) |
| chr2_+_208394794 | 1.96 |
ENST00000536726.1
ENST00000374397.4 ENST00000452474.1 |
CREB1
|
cAMP responsive element binding protein 1 |
| chr1_+_110453514 | 1.95 |
ENST00000369802.3
ENST00000420111.2 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr14_-_53417732 | 1.95 |
ENST00000399304.3
ENST00000395631.2 ENST00000341590.3 ENST00000343279.4 |
FERMT2
|
fermitin family member 2 |
| chr1_-_113249948 | 1.94 |
ENST00000339083.7
ENST00000369642.3 |
RHOC
|
ras homolog family member C |
| chr6_-_30710265 | 1.92 |
ENST00000438162.1
ENST00000454845.1 |
FLOT1
|
flotillin 1 |
| chr14_+_24616588 | 1.90 |
ENST00000324103.6
ENST00000559260.1 |
RNF31
|
ring finger protein 31 |
| chr6_+_31683117 | 1.86 |
ENST00000375825.3
ENST00000375824.1 |
LY6G6D
|
lymphocyte antigen 6 complex, locus G6D |
| chr7_-_102715263 | 1.83 |
ENST00000379305.3
|
FBXL13
|
F-box and leucine-rich repeat protein 13 |
| chr12_+_51632638 | 1.80 |
ENST00000549732.2
|
DAZAP2
|
DAZ associated protein 2 |
| chr2_+_208394658 | 1.72 |
ENST00000421139.1
|
CREB1
|
cAMP responsive element binding protein 1 |
| chrX_+_48755183 | 1.71 |
ENST00000376563.1
ENST00000376566.4 |
PQBP1
|
polyglutamine binding protein 1 |
| chr2_+_208394455 | 1.68 |
ENST00000430624.1
|
CREB1
|
cAMP responsive element binding protein 1 |
| chr15_-_89755034 | 1.64 |
ENST00000563254.1
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr10_+_89419370 | 1.62 |
ENST00000361175.4
ENST00000456849.1 |
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr14_-_24658053 | 1.59 |
ENST00000354464.6
|
IPO4
|
importin 4 |
| chr4_-_103749205 | 1.59 |
ENST00000508249.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr14_-_35873856 | 1.58 |
ENST00000553342.1
ENST00000216797.5 ENST00000557140.1 |
NFKBIA
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| chr3_+_101546827 | 1.56 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr16_+_84209738 | 1.53 |
ENST00000564928.1
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr1_-_27816641 | 1.52 |
ENST00000430629.2
|
WASF2
|
WAS protein family, member 2 |
| chr3_+_183967409 | 1.50 |
ENST00000324557.4
ENST00000402825.3 |
ECE2
|
endothelin converting enzyme 2 |
| chr16_-_2059748 | 1.50 |
ENST00000562103.1
ENST00000431526.1 |
ZNF598
|
zinc finger protein 598 |
| chr6_+_15401075 | 1.49 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr17_-_37382105 | 1.48 |
ENST00000333461.5
|
STAC2
|
SH3 and cysteine rich domain 2 |
| chr19_-_50370509 | 1.47 |
ENST00000596014.1
|
PNKP
|
polynucleotide kinase 3'-phosphatase |
| chr18_+_63418068 | 1.45 |
ENST00000397968.2
|
CDH7
|
cadherin 7, type 2 |
| chr13_-_38443860 | 1.45 |
ENST00000426868.2
ENST00000379681.3 ENST00000338947.5 ENST00000355779.2 ENST00000358477.2 ENST00000379673.2 |
TRPC4
|
transient receptor potential cation channel, subfamily C, member 4 |
| chr5_-_59064458 | 1.41 |
ENST00000502575.1
ENST00000507116.1 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr12_-_122985067 | 1.40 |
ENST00000540586.1
ENST00000543897.1 |
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr11_+_65292884 | 1.40 |
ENST00000527009.1
|
SCYL1
|
SCY1-like 1 (S. cerevisiae) |
| chr2_-_27357479 | 1.36 |
ENST00000406567.3
ENST00000260643.2 |
PREB
|
prolactin regulatory element binding |
| chr9_-_136004782 | 1.28 |
ENST00000393157.3
|
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr16_+_3014217 | 1.23 |
ENST00000572045.1
|
KREMEN2
|
kringle containing transmembrane protein 2 |
| chr4_+_169418195 | 1.23 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr3_-_58572760 | 1.15 |
ENST00000447756.2
|
FAM107A
|
family with sequence similarity 107, member A |
| chr20_-_33872518 | 1.14 |
ENST00000374436.3
|
EIF6
|
eukaryotic translation initiation factor 6 |
| chr3_+_42201653 | 1.14 |
ENST00000341421.3
ENST00000396175.1 |
TRAK1
|
trafficking protein, kinesin binding 1 |
| chrX_-_40594755 | 1.13 |
ENST00000324817.1
|
MED14
|
mediator complex subunit 14 |
| chr16_-_88717482 | 1.09 |
ENST00000261623.3
|
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr18_+_33877654 | 1.08 |
ENST00000257209.4
ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3
|
formin homology 2 domain containing 3 |
| chr22_-_24951054 | 1.07 |
ENST00000447813.2
ENST00000402766.1 ENST00000407471.3 ENST00000435822.1 |
GUCD1
|
guanylyl cyclase domain containing 1 |
| chr15_+_85923797 | 1.05 |
ENST00000559362.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr20_-_33872548 | 1.01 |
ENST00000374443.3
|
EIF6
|
eukaryotic translation initiation factor 6 |
| chr3_-_179322436 | 1.00 |
ENST00000392659.2
ENST00000476781.1 |
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr1_+_95582881 | 0.97 |
ENST00000370203.4
ENST00000456991.1 |
TMEM56
|
transmembrane protein 56 |
| chr2_-_18741882 | 0.95 |
ENST00000381249.3
|
RDH14
|
retinol dehydrogenase 14 (all-trans/9-cis/11-cis) |
| chr3_-_179322416 | 0.94 |
ENST00000259038.2
|
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr17_+_40985407 | 0.94 |
ENST00000586114.1
ENST00000590720.1 ENST00000585805.1 ENST00000541124.1 ENST00000441946.2 ENST00000591152.1 ENST00000589469.1 ENST00000293362.3 ENST00000592169.1 |
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki) |
| chr11_+_65479462 | 0.92 |
ENST00000377046.3
ENST00000352980.4 ENST00000341318.4 |
KAT5
|
K(lysine) acetyltransferase 5 |
| chr9_+_6328338 | 0.91 |
ENST00000344545.5
|
TPD52L3
|
tumor protein D52-like 3 |
| chr1_-_93426998 | 0.89 |
ENST00000370310.4
|
FAM69A
|
family with sequence similarity 69, member A |
| chr2_-_113594279 | 0.88 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr20_+_44486246 | 0.88 |
ENST00000255152.2
ENST00000454862.2 |
ZSWIM3
|
zinc finger, SWIM-type containing 3 |
| chr19_+_18794470 | 0.87 |
ENST00000321949.8
ENST00000338797.6 |
CRTC1
|
CREB regulated transcription coactivator 1 |
| chr4_+_169418255 | 0.87 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr3_+_52719936 | 0.87 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr12_+_51633061 | 0.85 |
ENST00000551313.1
|
DAZAP2
|
DAZ associated protein 2 |
| chr16_-_88717423 | 0.84 |
ENST00000568278.1
ENST00000569359.1 ENST00000567174.1 |
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr12_-_122985494 | 0.84 |
ENST00000336229.4
|
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr7_+_107384579 | 0.84 |
ENST00000222597.2
ENST00000415884.2 |
CBLL1
|
Cbl proto-oncogene-like 1, E3 ubiquitin protein ligase |
| chr17_-_4643114 | 0.82 |
ENST00000293778.6
|
CXCL16
|
chemokine (C-X-C motif) ligand 16 |
| chr4_-_176733897 | 0.81 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr16_+_67571351 | 0.81 |
ENST00000428437.2
ENST00000569253.1 |
FAM65A
|
family with sequence similarity 65, member A |
| chr4_-_103749179 | 0.81 |
ENST00000502690.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr4_-_103749105 | 0.81 |
ENST00000394801.4
ENST00000394804.2 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr17_+_34640031 | 0.81 |
ENST00000339270.6
ENST00000482104.1 |
CCL4L2
|
chemokine (C-C motif) ligand 4-like 2 |
| chr22_+_22676808 | 0.80 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr19_-_49250054 | 0.79 |
ENST00000602105.1
ENST00000332955.2 |
IZUMO1
|
izumo sperm-egg fusion 1 |
| chr10_+_70587279 | 0.79 |
ENST00000298596.6
ENST00000399169.4 ENST00000399165.4 ENST00000399162.2 |
STOX1
|
storkhead box 1 |
| chr15_+_26360970 | 0.78 |
ENST00000556159.1
ENST00000557523.1 |
LINC00929
|
long intergenic non-protein coding RNA 929 |
| chr11_-_104893863 | 0.78 |
ENST00000260315.3
ENST00000526056.1 ENST00000531367.1 ENST00000456094.1 ENST00000444749.2 ENST00000393141.2 ENST00000418434.1 ENST00000393139.2 |
CASP5
|
caspase 5, apoptosis-related cysteine peptidase |
| chr6_+_160327974 | 0.77 |
ENST00000252660.4
|
MAS1
|
MAS1 oncogene |
| chr10_-_29923893 | 0.77 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr3_-_178789993 | 0.77 |
ENST00000432729.1
|
ZMAT3
|
zinc finger, matrin-type 3 |
| chr4_-_54930790 | 0.76 |
ENST00000263921.3
|
CHIC2
|
cysteine-rich hydrophobic domain 2 |
| chr17_-_4852332 | 0.75 |
ENST00000572383.1
|
PFN1
|
profilin 1 |
| chr11_-_61734599 | 0.75 |
ENST00000532601.1
|
FTH1
|
ferritin, heavy polypeptide 1 |
| chr14_-_61190754 | 0.73 |
ENST00000216513.4
|
SIX4
|
SIX homeobox 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of REL
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 28.6 | GO:0052027 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 6.2 | 18.5 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 6.1 | 6.1 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 5.9 | 23.5 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 5.8 | 29.1 | GO:1903974 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 5.7 | 28.3 | GO:0070842 | aggresome assembly(GO:0070842) |
| 4.8 | 14.5 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 3.6 | 21.7 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 3.5 | 17.6 | GO:0033590 | response to cobalamin(GO:0033590) |
| 3.5 | 27.8 | GO:0021564 | vagus nerve development(GO:0021564) |
| 3.3 | 29.4 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 2.6 | 7.9 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 2.3 | 6.8 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 2.2 | 11.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 2.1 | 8.4 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 2.1 | 12.4 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 2.1 | 8.3 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 1.7 | 17.3 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 1.7 | 5.1 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 1.6 | 9.5 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 1.6 | 9.4 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 1.5 | 6.0 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 1.5 | 10.5 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 1.4 | 41.9 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 1.3 | 4.0 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 1.3 | 15.9 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 1.3 | 6.6 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 1.3 | 5.1 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 1.2 | 14.7 | GO:0032782 | bile acid secretion(GO:0032782) |
| 1.2 | 2.3 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 1.1 | 2.2 | GO:2000669 | negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 1.0 | 10.0 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 1.0 | 4.8 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.9 | 5.6 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.9 | 2.6 | GO:1904897 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.8 | 6.5 | GO:0030578 | PML body organization(GO:0030578) |
| 0.8 | 13.8 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.8 | 6.1 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.7 | 4.4 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.7 | 4.4 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.7 | 7.0 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.7 | 2.0 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.6 | 7.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.5 | 2.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.5 | 4.3 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.5 | 1.6 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.5 | 12.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.5 | 2.0 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.5 | 2.8 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.5 | 1.4 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.4 | 2.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.4 | 3.3 | GO:0051106 | positive regulation of DNA ligation(GO:0051106) |
| 0.4 | 1.6 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.4 | 12.9 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.4 | 3.5 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.4 | 1.9 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.4 | 2.3 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.4 | 1.5 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.3 | 2.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.3 | 1.9 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.3 | 2.5 | GO:0033504 | floor plate development(GO:0033504) |
| 0.3 | 1.5 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) DNA dephosphorylation(GO:0098502) |
| 0.3 | 0.9 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.2 | 0.7 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.2 | 1.9 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.2 | 4.9 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.2 | 7.8 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.2 | 0.7 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.2 | 2.0 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.2 | 0.6 | GO:1901189 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.2 | 2.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.2 | 5.8 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.2 | 0.7 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.2 | 4.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 1.6 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 1.4 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.2 | 4.3 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.2 | 11.5 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 2.9 | GO:0071786 | nuclear pore complex assembly(GO:0051292) endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.7 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.1 | 0.8 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 16.9 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.1 | 5.4 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 1.1 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.1 | 1.4 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 5.6 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.1 | 1.6 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 2.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 1.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 2.5 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.1 | 0.5 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 | 0.7 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.1 | 2.0 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.7 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 2.3 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.1 | 2.4 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.4 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.1 | 0.6 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.1 | 1.9 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 0.5 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.1 | 1.4 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 0.9 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 2.2 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 0.8 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 1.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.1 | GO:0045080 | positive regulation of chemokine biosynthetic process(GO:0045080) |
| 0.1 | 1.0 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.1 | 0.8 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 | 1.9 | GO:1905145 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 8.4 | GO:0048675 | axon extension(GO:0048675) |
| 0.1 | 0.8 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.1 | 0.3 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 3.9 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.1 | 0.2 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 0.5 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 4.5 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 4.4 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 2.4 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.9 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 1.1 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.3 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.1 | GO:0070563 | cell migration involved in endocardial cushion formation(GO:0003273) negative regulation of vitamin D receptor signaling pathway(GO:0070563) negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.8 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.4 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0036114 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.2 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.3 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.4 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.8 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.1 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.2 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 5.3 | GO:0045665 | negative regulation of neuron differentiation(GO:0045665) |
| 0.0 | 1.0 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 2.5 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 0.3 | GO:0010561 | negative regulation of glycoprotein biosynthetic process(GO:0010561) |
| 0.0 | 0.3 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.5 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.0 | 0.8 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 2.0 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 1.9 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 1.2 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.0 | 1.4 | GO:0060337 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 1.1 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.0 | GO:1904760 | peptidyl-serine ADP-ribosylation(GO:0018312) myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 | 0.2 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.0 | 1.5 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.8 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.2 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.0 | 1.2 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 3.0 | GO:0001666 | response to hypoxia(GO:0001666) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 23.5 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 4.8 | 29.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 2.2 | 17.6 | GO:0043196 | varicosity(GO:0043196) |
| 1.4 | 11.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 1.3 | 14.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 1.2 | 28.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 1.2 | 8.3 | GO:0042825 | TAP complex(GO:0042825) |
| 1.0 | 4.9 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 1.0 | 5.8 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.9 | 8.0 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.9 | 4.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.8 | 5.6 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.6 | 4.7 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.5 | 2.0 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.4 | 5.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.4 | 28.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.4 | 4.0 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.4 | 13.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.3 | 1.7 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.2 | 5.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.2 | 0.7 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.2 | 6.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 3.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 0.7 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.2 | 2.0 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 2.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 1.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.2 | 2.0 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 18.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 3.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.9 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 10.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 2.2 | GO:0033643 | host cell part(GO:0033643) |
| 0.1 | 2.0 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 0.9 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.7 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 0.8 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.1 | 3.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.6 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 2.4 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 5.8 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 2.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 37.5 | GO:0045121 | membrane raft(GO:0045121) |
| 0.1 | 3.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 1.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.6 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 5.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 2.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.6 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 2.0 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.1 | 9.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.5 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 8.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 0.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 99.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.8 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 2.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 1.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 3.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.8 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 19.4 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 25.4 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.4 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 1.1 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 1.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.9 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 5.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 15.1 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 4.2 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 1.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.1 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 2.4 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 1.1 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 6.2 | GO:0044297 | cell body(GO:0044297) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 14.5 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 3.8 | 60.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 3.6 | 29.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 3.4 | 16.9 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 2.3 | 7.0 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 2.2 | 11.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 1.9 | 9.5 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 1.6 | 9.4 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 1.3 | 28.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 1.2 | 8.3 | GO:0046979 | TAP2 binding(GO:0046979) |
| 1.2 | 18.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 1.1 | 36.9 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 1.0 | 4.0 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 1.0 | 3.0 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) |
| 0.9 | 13.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.8 | 5.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.8 | 4.9 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.8 | 2.3 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.7 | 5.6 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.7 | 36.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.7 | 29.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.6 | 1.9 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.5 | 4.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.5 | 1.6 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.5 | 7.8 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.5 | 4.8 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.5 | 5.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.4 | 7.9 | GO:0048185 | activin binding(GO:0048185) |
| 0.4 | 3.9 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.4 | 6.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.4 | 1.5 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) polynucleotide phosphatase activity(GO:0098518) |
| 0.3 | 16.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.3 | 8.0 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 1.6 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.2 | 2.6 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.2 | 4.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.2 | 2.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.2 | 2.3 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.2 | 2.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 2.0 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.2 | 2.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 0.7 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 3.1 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 3.3 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 5.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 31.0 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 2.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 4.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.7 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 2.0 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.6 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 0.8 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 3.0 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 2.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 9.6 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 0.5 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.1 | 1.4 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 1.9 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 1.0 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 4.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.7 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 3.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 1.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 2.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 13.8 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 1.4 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 2.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 2.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 1.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.3 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 23.1 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.1 | 8.6 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 0.9 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 2.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.5 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 5.5 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.8 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 2.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 3.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 5.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.9 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.9 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 1.9 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 3.5 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.9 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 1.5 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 2.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.6 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.0 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 11.9 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 88.8 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 1.0 | 30.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.7 | 30.8 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.5 | 9.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.4 | 4.9 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.4 | 7.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.4 | 0.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.3 | 9.7 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.3 | 24.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.3 | 9.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.3 | 8.4 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.3 | 8.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.3 | 23.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.2 | 4.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 7.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 13.1 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.2 | 6.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.2 | 68.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.2 | 4.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 6.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 10.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 4.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 4.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 4.5 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 6.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 2.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 6.8 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 3.3 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 1.9 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 1.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 2.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.4 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 3.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 4.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 1.4 | 31.0 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 1.1 | 75.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 1.0 | 53.0 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.6 | 13.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.6 | 5.1 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.5 | 19.1 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.4 | 13.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.4 | 7.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 8.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.3 | 10.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.3 | 6.6 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.2 | 18.9 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.2 | 5.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 5.6 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.2 | 7.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 17.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 4.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 4.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 6.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 4.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.4 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 5.1 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 4.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 7.6 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 1.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 8.4 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 1.9 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.1 | 1.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 2.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 2.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.7 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 1.4 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.1 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |