Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
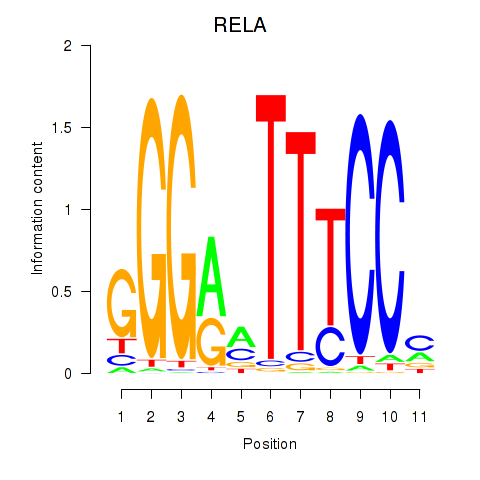
Results for RELA
Z-value: 3.69
Transcription factors associated with RELA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RELA
|
ENSG00000173039.14 | RELA proto-oncogene, NF-kB subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RELA | hg19_v2_chr11_-_65430251_65430399 | 0.94 | 5.1e-12 | Click! |
Activity profile of RELA motif
Sorted Z-values of RELA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_76944621 | 64.70 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr6_-_31550192 | 45.65 |
ENST00000429299.2
ENST00000446745.2 |
LTB
|
lymphotoxin beta (TNF superfamily, member 3) |
| chr6_-_29527702 | 38.52 |
ENST00000377050.4
|
UBD
|
ubiquitin D |
| chr19_+_45504688 | 35.86 |
ENST00000221452.8
ENST00000540120.1 ENST00000505236.1 |
RELB
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr11_+_102188224 | 33.63 |
ENST00000263464.3
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr6_+_138188551 | 32.02 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr11_+_102188272 | 31.75 |
ENST00000532808.1
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr10_+_104154229 | 30.96 |
ENST00000428099.1
ENST00000369966.3 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr4_+_74702214 | 30.75 |
ENST00000226317.5
ENST00000515050.1 |
CXCL6
|
chemokine (C-X-C motif) ligand 6 |
| chr2_+_228678550 | 29.39 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr5_-_150460914 | 27.29 |
ENST00000389378.2
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr19_+_10381769 | 25.48 |
ENST00000423829.2
ENST00000588645.1 |
ICAM1
|
intercellular adhesion molecule 1 |
| chr4_-_76957214 | 24.75 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr4_+_103422471 | 24.46 |
ENST00000226574.4
ENST00000394820.4 |
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr1_+_101185290 | 23.98 |
ENST00000370119.4
ENST00000347652.2 ENST00000294728.2 ENST00000370115.1 |
VCAM1
|
vascular cell adhesion molecule 1 |
| chr1_+_110453109 | 21.86 |
ENST00000525659.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr4_-_74864386 | 21.60 |
ENST00000296027.4
|
CXCL5
|
chemokine (C-X-C motif) ligand 5 |
| chr4_-_74904398 | 20.44 |
ENST00000296026.4
|
CXCL3
|
chemokine (C-X-C motif) ligand 3 |
| chr1_-_8000872 | 19.27 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr5_-_150460539 | 19.23 |
ENST00000520931.1
ENST00000520695.1 ENST00000521591.1 ENST00000518977.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr14_+_103589789 | 19.18 |
ENST00000558056.1
ENST00000560869.1 |
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr11_+_19798964 | 19.08 |
ENST00000527559.2
|
NAV2
|
neuron navigator 2 |
| chr1_+_110453203 | 19.00 |
ENST00000357302.4
ENST00000344188.5 ENST00000329608.6 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr4_-_74964904 | 18.68 |
ENST00000508487.2
|
CXCL2
|
chemokine (C-X-C motif) ligand 2 |
| chr4_+_74735102 | 18.52 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr19_+_4229495 | 18.18 |
ENST00000221847.5
|
EBI3
|
Epstein-Barr virus induced 3 |
| chr12_-_49259643 | 17.76 |
ENST00000309739.5
|
RND1
|
Rho family GTPase 1 |
| chr11_+_19799327 | 17.72 |
ENST00000540292.1
|
NAV2
|
neuron navigator 2 |
| chr12_-_9913489 | 17.33 |
ENST00000228434.3
ENST00000536709.1 |
CD69
|
CD69 molecule |
| chr14_-_35873856 | 17.26 |
ENST00000553342.1
ENST00000216797.5 ENST00000557140.1 |
NFKBIA
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| chr5_-_150466692 | 17.09 |
ENST00000315050.7
ENST00000523338.1 ENST00000522100.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr12_-_57504069 | 16.74 |
ENST00000543873.2
ENST00000554663.1 ENST00000557635.1 |
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr10_+_104155450 | 15.78 |
ENST00000471698.1
ENST00000189444.6 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr16_+_50776021 | 15.65 |
ENST00000566679.2
ENST00000564634.1 ENST00000398568.2 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr1_+_110453462 | 14.82 |
ENST00000488198.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr3_+_53195136 | 13.90 |
ENST00000394729.2
ENST00000330452.3 |
PRKCD
|
protein kinase C, delta |
| chr17_+_40440481 | 13.35 |
ENST00000590726.2
ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A
|
signal transducer and activator of transcription 5A |
| chr1_-_209825674 | 12.66 |
ENST00000367030.3
ENST00000356082.4 |
LAMB3
|
laminin, beta 3 |
| chr16_+_50775948 | 12.57 |
ENST00000569681.1
ENST00000569418.1 ENST00000540145.1 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr16_+_50775971 | 12.45 |
ENST00000311559.9
ENST00000564326.1 ENST00000566206.1 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr4_-_120550146 | 11.62 |
ENST00000354960.3
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific |
| chr20_+_44746885 | 11.51 |
ENST00000372285.3
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chr11_-_8832182 | 11.50 |
ENST00000527510.1
ENST00000528527.1 ENST00000528523.1 ENST00000313726.6 |
ST5
|
suppression of tumorigenicity 5 |
| chr2_+_161993465 | 11.21 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr9_-_136344197 | 11.00 |
ENST00000414172.1
ENST00000371897.4 |
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr19_-_47734448 | 9.72 |
ENST00000439096.2
|
BBC3
|
BCL2 binding component 3 |
| chr19_-_4338783 | 9.45 |
ENST00000601482.1
ENST00000600324.1 |
STAP2
|
signal transducing adaptor family member 2 |
| chr19_-_4338838 | 9.40 |
ENST00000594605.1
|
STAP2
|
signal transducing adaptor family member 2 |
| chr17_-_34207295 | 9.29 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr11_-_128392085 | 9.28 |
ENST00000526145.2
ENST00000531611.1 ENST00000319397.6 ENST00000345075.4 ENST00000535549.1 |
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr5_+_159895275 | 9.25 |
ENST00000517927.1
|
MIR146A
|
microRNA 146a |
| chr21_+_34775698 | 9.06 |
ENST00000381995.1
|
IFNGR2
|
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr18_-_71959159 | 9.02 |
ENST00000494131.2
ENST00000397914.4 ENST00000340533.4 |
CYB5A
|
cytochrome b5 type A (microsomal) |
| chr21_+_34775772 | 8.82 |
ENST00000405436.1
|
IFNGR2
|
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr21_+_34775181 | 8.69 |
ENST00000290219.6
|
IFNGR2
|
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr19_+_45251804 | 8.66 |
ENST00000164227.5
|
BCL3
|
B-cell CLL/lymphoma 3 |
| chr7_-_47579188 | 8.55 |
ENST00000398879.1
ENST00000355730.3 ENST00000442536.2 ENST00000458317.2 |
TNS3
|
tensin 3 |
| chr17_-_61777090 | 8.49 |
ENST00000578061.1
|
LIMD2
|
LIM domain containing 2 |
| chr2_-_163175133 | 8.44 |
ENST00000421365.2
ENST00000263642.2 |
IFIH1
|
interferon induced with helicase C domain 1 |
| chr14_+_103243813 | 8.43 |
ENST00000560371.1
ENST00000347662.4 ENST00000392745.2 ENST00000539721.1 ENST00000560463.1 |
TRAF3
|
TNF receptor-associated factor 3 |
| chr22_-_19512893 | 8.17 |
ENST00000403084.1
ENST00000413119.2 |
CLDN5
|
claudin 5 |
| chr8_+_86157699 | 8.13 |
ENST00000321764.3
|
CA13
|
carbonic anhydrase XIII |
| chr7_-_22396533 | 8.02 |
ENST00000344041.6
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr9_-_117568365 | 7.88 |
ENST00000374045.4
|
TNFSF15
|
tumor necrosis factor (ligand) superfamily, member 15 |
| chr2_+_161993412 | 7.86 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr11_-_72492903 | 7.81 |
ENST00000537947.1
|
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr16_+_50730910 | 7.62 |
ENST00000300589.2
|
NOD2
|
nucleotide-binding oligomerization domain containing 2 |
| chr17_+_21191341 | 7.51 |
ENST00000526076.2
ENST00000361818.5 ENST00000316920.6 |
MAP2K3
|
mitogen-activated protein kinase kinase 3 |
| chr20_-_43977055 | 7.46 |
ENST00000372733.3
ENST00000537976.1 |
SDC4
|
syndecan 4 |
| chr3_+_101546827 | 7.36 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr4_-_185395672 | 7.33 |
ENST00000393593.3
|
IRF2
|
interferon regulatory factor 2 |
| chr9_-_32526184 | 7.27 |
ENST00000545044.1
ENST00000379868.1 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr19_-_17958832 | 7.24 |
ENST00000458235.1
|
JAK3
|
Janus kinase 3 |
| chr10_+_30722866 | 7.16 |
ENST00000263056.1
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr14_+_73704201 | 7.16 |
ENST00000340738.5
ENST00000427855.1 ENST00000381166.3 |
PAPLN
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr11_-_72492878 | 6.88 |
ENST00000535054.1
ENST00000545082.1 |
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr10_+_13142075 | 6.70 |
ENST00000378757.2
ENST00000430081.1 ENST00000378752.3 ENST00000378748.3 |
OPTN
|
optineurin |
| chr10_+_13142225 | 6.53 |
ENST00000378747.3
|
OPTN
|
optineurin |
| chr4_-_122085469 | 6.50 |
ENST00000057513.3
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr1_+_169077172 | 6.48 |
ENST00000499679.3
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr13_+_97874574 | 6.19 |
ENST00000343600.4
ENST00000345429.6 ENST00000376673.3 |
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr14_+_22984601 | 6.14 |
ENST00000390509.1
|
TRAJ28
|
T cell receptor alpha joining 28 |
| chr10_+_13141585 | 6.06 |
ENST00000378764.2
|
OPTN
|
optineurin |
| chr3_-_4793274 | 5.81 |
ENST00000414938.1
|
EGOT
|
eosinophil granule ontogeny transcript (non-protein coding) |
| chr6_+_29691198 | 5.60 |
ENST00000440587.2
ENST00000434407.2 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr1_+_100818156 | 5.59 |
ENST00000336454.3
|
CDC14A
|
cell division cycle 14A |
| chr15_+_52311398 | 5.55 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr14_-_24616426 | 5.52 |
ENST00000216802.5
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr1_-_209824643 | 5.43 |
ENST00000391911.1
ENST00000415782.1 |
LAMB3
|
laminin, beta 3 |
| chr14_+_93897272 | 5.30 |
ENST00000393151.2
|
UNC79
|
unc-79 homolog (C. elegans) |
| chr1_-_65432171 | 5.28 |
ENST00000342505.4
|
JAK1
|
Janus kinase 1 |
| chr2_+_97202480 | 5.26 |
ENST00000357485.3
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr2_+_97203082 | 5.11 |
ENST00000454558.2
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr19_+_39390320 | 5.07 |
ENST00000576510.1
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr7_+_143078652 | 5.06 |
ENST00000354434.4
ENST00000449423.2 |
ZYX
|
zyxin |
| chr6_+_29691056 | 5.05 |
ENST00000414333.1
ENST00000334668.4 ENST00000259951.7 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr16_+_53738053 | 4.85 |
ENST00000394647.3
|
FTO
|
fat mass and obesity associated |
| chr11_-_75062829 | 4.82 |
ENST00000393505.4
|
ARRB1
|
arrestin, beta 1 |
| chr6_+_135502466 | 4.78 |
ENST00000367814.4
|
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr11_-_3862206 | 4.70 |
ENST00000351018.4
|
RHOG
|
ras homolog family member G |
| chr1_+_6845384 | 4.53 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr2_+_162016916 | 4.53 |
ENST00000405852.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr10_+_12391481 | 4.51 |
ENST00000378847.3
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID |
| chr5_+_14143728 | 4.51 |
ENST00000344204.4
ENST00000537187.1 |
TRIO
|
trio Rho guanine nucleotide exchange factor |
| chr7_-_98741642 | 4.40 |
ENST00000361368.2
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1 |
| chr19_+_39390587 | 4.35 |
ENST00000572515.1
ENST00000392079.3 ENST00000575359.1 ENST00000313582.5 |
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr12_-_323689 | 4.23 |
ENST00000428720.1
|
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr7_-_98741714 | 4.23 |
ENST00000361125.1
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1 |
| chr4_+_4861385 | 4.22 |
ENST00000382723.4
|
MSX1
|
msh homeobox 1 |
| chr11_-_75062730 | 4.16 |
ENST00000420843.2
ENST00000360025.3 |
ARRB1
|
arrestin, beta 1 |
| chr1_+_100818009 | 4.15 |
ENST00000370125.2
ENST00000361544.6 ENST00000370124.3 |
CDC14A
|
cell division cycle 14A |
| chr3_-_58563094 | 4.09 |
ENST00000464064.1
|
FAM107A
|
family with sequence similarity 107, member A |
| chr14_+_93897199 | 3.89 |
ENST00000553484.1
|
UNC79
|
unc-79 homolog (C. elegans) |
| chr6_+_144471643 | 3.88 |
ENST00000367568.4
|
STX11
|
syntaxin 11 |
| chr15_+_44719970 | 3.86 |
ENST00000558966.1
|
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr1_+_110453514 | 3.81 |
ENST00000369802.3
ENST00000420111.2 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr5_+_133984462 | 3.78 |
ENST00000398844.2
ENST00000322887.4 |
SEC24A
|
SEC24 family member A |
| chr14_-_53417732 | 3.73 |
ENST00000399304.3
ENST00000395631.2 ENST00000341590.3 ENST00000343279.4 |
FERMT2
|
fermitin family member 2 |
| chr3_+_9691117 | 3.62 |
ENST00000353332.5
ENST00000420925.1 ENST00000296003.4 ENST00000351233.5 |
MTMR14
|
myotubularin related protein 14 |
| chr6_+_106534192 | 3.52 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr19_+_41725140 | 3.42 |
ENST00000359092.3
|
AXL
|
AXL receptor tyrosine kinase |
| chr20_+_18488528 | 3.36 |
ENST00000377465.1
|
SEC23B
|
Sec23 homolog B (S. cerevisiae) |
| chr2_-_209028300 | 3.34 |
ENST00000304502.4
|
CRYGA
|
crystallin, gamma A |
| chr4_-_103749205 | 3.31 |
ENST00000508249.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr2_-_136743039 | 3.28 |
ENST00000537273.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr17_-_7590745 | 3.26 |
ENST00000514944.1
ENST00000503591.1 ENST00000455263.2 ENST00000420246.2 ENST00000445888.2 ENST00000509690.1 ENST00000604348.1 ENST00000269305.4 |
TP53
|
tumor protein p53 |
| chr9_+_132934835 | 3.22 |
ENST00000372398.3
|
NCS1
|
neuronal calcium sensor 1 |
| chr2_+_208394455 | 3.22 |
ENST00000430624.1
|
CREB1
|
cAMP responsive element binding protein 1 |
| chr4_-_76928641 | 3.15 |
ENST00000264888.5
|
CXCL9
|
chemokine (C-X-C motif) ligand 9 |
| chr20_+_18488137 | 3.11 |
ENST00000450074.1
ENST00000262544.2 ENST00000336714.3 ENST00000377475.3 |
SEC23B
|
Sec23 homolog B (S. cerevisiae) |
| chr2_+_208394794 | 3.07 |
ENST00000536726.1
ENST00000374397.4 ENST00000452474.1 |
CREB1
|
cAMP responsive element binding protein 1 |
| chr11_+_118478313 | 3.07 |
ENST00000356063.5
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1 |
| chr2_-_136743169 | 3.06 |
ENST00000264161.4
|
DARS
|
aspartyl-tRNA synthetase |
| chr6_+_29910301 | 3.06 |
ENST00000376809.5
ENST00000376802.2 |
HLA-A
|
major histocompatibility complex, class I, A |
| chr1_+_44870866 | 3.01 |
ENST00000355387.2
ENST00000361799.2 |
RNF220
|
ring finger protein 220 |
| chr1_+_100818484 | 2.96 |
ENST00000544534.1
|
CDC14A
|
cell division cycle 14A |
| chr22_-_38380543 | 2.96 |
ENST00000396884.2
|
SOX10
|
SRY (sex determining region Y)-box 10 |
| chr12_+_49761147 | 2.90 |
ENST00000549298.1
|
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr21_-_43373999 | 2.87 |
ENST00000380486.3
|
C2CD2
|
C2 calcium-dependent domain containing 2 |
| chr4_+_146560245 | 2.74 |
ENST00000541599.1
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr12_+_53662073 | 2.69 |
ENST00000553219.1
ENST00000257934.4 |
ESPL1
|
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr2_+_208394658 | 2.67 |
ENST00000421139.1
|
CREB1
|
cAMP responsive element binding protein 1 |
| chr5_-_132113036 | 2.66 |
ENST00000378706.1
|
SEPT8
|
septin 8 |
| chr9_-_32526299 | 2.64 |
ENST00000379882.1
ENST00000379883.2 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr11_+_6625046 | 2.63 |
ENST00000396751.2
|
ILK
|
integrin-linked kinase |
| chrX_-_48901012 | 2.59 |
ENST00000315869.7
|
TFE3
|
transcription factor binding to IGHM enhancer 3 |
| chr16_-_66959429 | 2.57 |
ENST00000420652.1
ENST00000299759.6 |
RRAD
|
Ras-related associated with diabetes |
| chr11_+_6624955 | 2.57 |
ENST00000299421.4
ENST00000537806.1 |
ILK
|
integrin-linked kinase |
| chr20_-_62203808 | 2.55 |
ENST00000467148.1
|
HELZ2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr1_-_155947951 | 2.54 |
ENST00000313695.7
ENST00000497907.1 |
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr5_-_132112907 | 2.52 |
ENST00000458488.2
|
SEPT8
|
septin 8 |
| chr12_+_53662110 | 2.51 |
ENST00000552462.1
|
ESPL1
|
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr13_-_38443860 | 2.50 |
ENST00000426868.2
ENST00000379681.3 ENST00000338947.5 ENST00000355779.2 ENST00000358477.2 ENST00000379673.2 |
TRPC4
|
transient receptor potential cation channel, subfamily C, member 4 |
| chr5_-_132112921 | 2.49 |
ENST00000378721.4
ENST00000378701.1 |
SEPT8
|
septin 8 |
| chr1_-_1822495 | 2.46 |
ENST00000378609.4
|
GNB1
|
guanine nucleotide binding protein (G protein), beta polypeptide 1 |
| chr2_+_64681641 | 2.45 |
ENST00000409537.2
|
LGALSL
|
lectin, galactoside-binding-like |
| chr22_-_28315115 | 2.43 |
ENST00000455418.3
ENST00000436663.1 ENST00000320996.10 ENST00000335272.5 |
PITPNB
|
phosphatidylinositol transfer protein, beta |
| chr15_+_85923797 | 2.40 |
ENST00000559362.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr5_-_58652788 | 2.35 |
ENST00000405755.2
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr20_+_34359905 | 2.30 |
ENST00000374012.3
ENST00000439301.1 ENST00000339089.6 ENST00000374000.4 |
PHF20
|
PHD finger protein 20 |
| chr2_+_124782857 | 2.27 |
ENST00000431078.1
|
CNTNAP5
|
contactin associated protein-like 5 |
| chr4_+_140222609 | 2.23 |
ENST00000296543.5
ENST00000398947.1 |
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr19_+_51728316 | 2.19 |
ENST00000436584.2
ENST00000421133.2 ENST00000391796.3 ENST00000262262.4 |
CD33
|
CD33 molecule |
| chr20_-_4795747 | 2.19 |
ENST00000379376.2
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2 |
| chr5_-_58335281 | 2.17 |
ENST00000358923.6
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr19_+_496454 | 2.16 |
ENST00000346144.4
ENST00000215637.3 ENST00000382683.4 |
MADCAM1
|
mucosal vascular addressin cell adhesion molecule 1 |
| chrX_-_131352152 | 2.12 |
ENST00000342983.2
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr18_-_12884259 | 2.11 |
ENST00000353319.4
ENST00000327283.3 |
PTPN2
|
protein tyrosine phosphatase, non-receptor type 2 |
| chr5_-_132113083 | 2.10 |
ENST00000296873.7
|
SEPT8
|
septin 8 |
| chr9_+_36572851 | 2.10 |
ENST00000298048.2
ENST00000538311.1 ENST00000536987.1 ENST00000545008.1 ENST00000536860.1 ENST00000536329.1 ENST00000541717.1 ENST00000543751.1 |
MELK
|
maternal embryonic leucine zipper kinase |
| chr17_+_16318850 | 2.09 |
ENST00000338560.7
|
TRPV2
|
transient receptor potential cation channel, subfamily V, member 2 |
| chr17_+_16318909 | 2.07 |
ENST00000577397.1
|
TRPV2
|
transient receptor potential cation channel, subfamily V, member 2 |
| chr3_+_157154578 | 2.06 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr11_+_94501497 | 2.05 |
ENST00000317829.8
ENST00000317837.9 ENST00000433060.2 |
AMOTL1
|
angiomotin like 1 |
| chr7_+_134464376 | 2.05 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr6_-_136847099 | 2.01 |
ENST00000438100.2
|
MAP7
|
microtubule-associated protein 7 |
| chr1_+_107683436 | 2.01 |
ENST00000370068.1
|
NTNG1
|
netrin G1 |
| chr10_-_43904235 | 1.98 |
ENST00000356053.3
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr12_+_49761224 | 1.98 |
ENST00000553127.1
ENST00000321898.6 |
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr1_+_151171012 | 1.96 |
ENST00000349792.5
ENST00000409426.1 ENST00000441902.2 ENST00000368890.4 ENST00000424999.1 ENST00000368888.4 |
PIP5K1A
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha |
| chr11_+_65292884 | 1.96 |
ENST00000527009.1
|
SCYL1
|
SCY1-like 1 (S. cerevisiae) |
| chr1_+_107683644 | 1.92 |
ENST00000370067.1
|
NTNG1
|
netrin G1 |
| chr10_+_86088381 | 1.91 |
ENST00000224756.8
ENST00000372088.2 |
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr12_-_48499591 | 1.90 |
ENST00000551330.1
ENST00000004980.5 ENST00000339976.6 ENST00000448372.1 |
SENP1
|
SUMO1/sentrin specific peptidase 1 |
| chr16_+_67571351 | 1.90 |
ENST00000428437.2
ENST00000569253.1 |
FAM65A
|
family with sequence similarity 65, member A |
| chr3_+_184032919 | 1.87 |
ENST00000427845.1
ENST00000342981.4 ENST00000319274.6 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr20_+_1115821 | 1.82 |
ENST00000435720.1
|
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr22_-_37640456 | 1.80 |
ENST00000405484.1
ENST00000441619.1 ENST00000406508.1 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr4_-_103749105 | 1.76 |
ENST00000394801.4
ENST00000394804.2 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr3_+_184033135 | 1.74 |
ENST00000424196.1
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr1_-_46664074 | 1.74 |
ENST00000371986.3
ENST00000371984.3 |
POMGNT1
|
protein O-linked mannose N-acetylglucosaminyltransferase 1 (beta 1,2-) |
| chr20_-_2451395 | 1.72 |
ENST00000339610.6
ENST00000381342.2 ENST00000438552.2 |
SNRPB
|
small nuclear ribonucleoprotein polypeptides B and B1 |
| chr15_+_26360970 | 1.71 |
ENST00000556159.1
ENST00000557523.1 |
LINC00929
|
long intergenic non-protein coding RNA 929 |
| chr19_+_41281060 | 1.70 |
ENST00000594436.1
ENST00000597784.1 |
MIA
|
melanoma inhibitory activity |
| chr12_-_6716569 | 1.66 |
ENST00000544040.1
ENST00000545942.1 |
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr1_-_39395165 | 1.65 |
ENST00000372985.3
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr12_-_6716534 | 1.61 |
ENST00000544484.1
ENST00000309577.6 ENST00000357008.2 |
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr14_+_24616588 | 1.61 |
ENST00000324103.6
ENST00000559260.1 |
RNF31
|
ring finger protein 31 |
| chr18_+_63418068 | 1.61 |
ENST00000397968.2
|
CDH7
|
cadherin 7, type 2 |
| chr19_-_4831701 | 1.59 |
ENST00000248244.5
|
TICAM1
|
toll-like receptor adaptor molecule 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RELA
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.9 | 63.6 | GO:0085032 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 13.9 | 55.4 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 13.6 | 40.7 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 11.9 | 59.5 | GO:1903974 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 10.7 | 32.0 | GO:0034146 | B-1 B cell homeostasis(GO:0001922) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 9.8 | 29.4 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 9.3 | 92.6 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 7.8 | 70.1 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 7.7 | 38.5 | GO:0070842 | aggresome assembly(GO:0070842) |
| 7.6 | 7.6 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 7.3 | 65.4 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 7.1 | 49.5 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 5.8 | 17.3 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 4.6 | 36.8 | GO:0021564 | vagus nerve development(GO:0021564) |
| 4.6 | 18.3 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 4.2 | 16.7 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 3.9 | 114.1 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 3.9 | 23.6 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 3.5 | 13.9 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 3.3 | 13.3 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 3.2 | 19.3 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 3.1 | 27.9 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 2.3 | 9.3 | GO:2000110 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) negative regulation of macrophage apoptotic process(GO:2000110) |
| 2.3 | 11.5 | GO:0033590 | response to cobalamin(GO:0033590) |
| 2.3 | 6.9 | GO:0002730 | regulation of dendritic cell cytokine production(GO:0002730) negative regulation of dendritic cell cytokine production(GO:0002731) |
| 2.2 | 28.7 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 2.1 | 6.3 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 1.7 | 18.2 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 1.6 | 3.3 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 1.6 | 4.8 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 1.6 | 4.8 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 1.6 | 9.3 | GO:0034670 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 1.5 | 3.1 | GO:2000669 | negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 1.5 | 7.5 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 1.5 | 10.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 1.4 | 4.2 | GO:0090427 | activation of meiosis(GO:0090427) |
| 1.4 | 8.4 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 1.4 | 13.7 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 1.3 | 6.5 | GO:1903281 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 1.2 | 14.7 | GO:0032782 | bile acid secretion(GO:0032782) |
| 1.2 | 9.3 | GO:0030578 | PML body organization(GO:0030578) |
| 1.1 | 17.8 | GO:0016322 | neuron remodeling(GO:0016322) |
| 1.1 | 19.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 1.1 | 3.2 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 1.0 | 8.2 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.9 | 7.5 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.9 | 3.7 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.9 | 9.0 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.8 | 1.6 | GO:0045350 | interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.8 | 12.7 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.8 | 3.8 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.8 | 27.1 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.8 | 4.5 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.7 | 3.0 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.7 | 3.5 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.7 | 2.7 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.6 | 2.6 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.6 | 3.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.6 | 18.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.5 | 9.2 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.5 | 4.5 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.4 | 1.3 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.4 | 9.0 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.4 | 3.3 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.4 | 8.9 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.4 | 1.4 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.3 | 2.1 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.3 | 11.0 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.3 | 1.0 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.3 | 6.7 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.3 | 2.0 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.3 | 1.0 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.3 | 1.6 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.3 | 2.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.3 | 13.9 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.3 | 3.6 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.3 | 0.8 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.2 | 1.9 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.2 | 0.7 | GO:1904529 | regulation of actin filament binding(GO:1904529) negative regulation of actin filament binding(GO:1904530) regulation of actin binding(GO:1904616) negative regulation of actin binding(GO:1904617) |
| 0.2 | 9.7 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.2 | 2.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.2 | 1.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.2 | 0.6 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.2 | 0.6 | GO:0031938 | regulation of chromatin silencing at telomere(GO:0031938) |
| 0.2 | 5.2 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.2 | 3.9 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.2 | 2.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.2 | 12.3 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.2 | 1.0 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.2 | 5.9 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.2 | 3.7 | GO:0033622 | integrin activation(GO:0033622) |
| 0.2 | 2.5 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.2 | 0.5 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 4.4 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 8.1 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 1.2 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.1 | 12.5 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.1 | 1.5 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 2.3 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 2.0 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 2.5 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 2.0 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.1 | 1.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 2.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 7.5 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) response to interleukin-12(GO:0070671) cellular response to interleukin-12(GO:0071349) |
| 0.1 | 0.3 | GO:1902218 | regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 | 0.5 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 6.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.4 | GO:1900194 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 2.2 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.1 | 0.8 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 5.5 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.1 | 5.7 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 4.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 1.6 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 1.4 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 | 0.5 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.9 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 1.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 7.2 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 3.0 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 0.6 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 1.1 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.1 | 0.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.3 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.1 | 2.1 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.4 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 5.1 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 7.4 | GO:0071222 | cellular response to lipopolysaccharide(GO:0071222) |
| 0.0 | 0.3 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 1.7 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.0 | 1.3 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 3.9 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.5 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 3.1 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 4.2 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.0 | 0.1 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.0 | 0.5 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) |
| 0.0 | 1.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 1.9 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 1.8 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.0 | 2.0 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.3 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 0.0 | 1.4 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.2 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.3 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 1.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.6 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.9 | 55.4 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 9.9 | 59.5 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 9.0 | 72.0 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 8.0 | 24.0 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 2.3 | 18.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 1.5 | 36.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 1.4 | 11.5 | GO:0043196 | varicosity(GO:0043196) |
| 1.3 | 14.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 1.3 | 9.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 1.2 | 13.7 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 1.2 | 14.7 | GO:0060091 | kinocilium(GO:0060091) |
| 0.9 | 5.5 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.8 | 4.8 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.7 | 40.7 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.7 | 8.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.6 | 38.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.5 | 19.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.4 | 10.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.4 | 25.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.3 | 6.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.3 | 21.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.3 | 6.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.3 | 8.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.3 | 2.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.3 | 1.4 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.3 | 8.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.3 | 5.5 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.3 | 3.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 12.5 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 3.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 3.1 | GO:0033643 | host cell part(GO:0033643) |
| 0.2 | 82.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.2 | 78.5 | GO:0045121 | membrane raft(GO:0045121) |
| 0.2 | 0.6 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.2 | 0.6 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.2 | 0.6 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.2 | 1.7 | GO:0071204 | U7 snRNP(GO:0005683) histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.2 | 3.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 3.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 2.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 1.6 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 6.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 2.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 2.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 11.6 | GO:0035580 | specific granule lumen(GO:0035580) tertiary granule lumen(GO:1904724) |
| 0.1 | 1.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 3.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 2.0 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 6.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 4.7 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.5 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 176.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 9.0 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 3.0 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.1 | 4.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 5.5 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 49.3 | GO:0005912 | adherens junction(GO:0005912) |
| 0.1 | 3.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 3.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 2.3 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 1.8 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 2.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 6.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 28.0 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 9.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 5.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 3.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 25.0 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 0.9 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 2.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 1.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 2.2 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.6 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 2.0 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 18.5 | 92.6 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 10.0 | 110.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 9.8 | 29.4 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 7.4 | 59.5 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 5.3 | 26.6 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 4.5 | 72.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 3.9 | 23.6 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 3.0 | 24.0 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 3.0 | 9.0 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 1.9 | 40.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 1.7 | 89.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 1.6 | 4.8 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 1.5 | 10.6 | GO:0046979 | TAP2 binding(GO:0046979) |
| 1.5 | 14.6 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 1.4 | 62.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 1.3 | 6.3 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 1.2 | 5.9 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 1.2 | 9.3 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.9 | 46.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.8 | 30.8 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.8 | 5.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.7 | 7.5 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.7 | 11.0 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.7 | 2.2 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.7 | 4.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.7 | 2.0 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.6 | 8.0 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.6 | 2.5 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.6 | 1.7 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.5 | 2.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.5 | 8.6 | GO:0048185 | activin binding(GO:0048185) |
| 0.5 | 1.4 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.5 | 5.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.5 | 8.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.4 | 17.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.4 | 4.8 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.4 | 11.6 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.4 | 6.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.3 | 18.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.3 | 10.4 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.3 | 6.5 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.3 | 1.9 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.3 | 3.3 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.3 | 1.7 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.3 | 3.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.3 | 9.0 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 2.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.3 | 28.6 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.3 | 7.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.2 | 55.9 | GO:0004386 | helicase activity(GO:0004386) |
| 0.2 | 7.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 4.5 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.2 | 1.3 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.2 | 3.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 7.9 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.2 | 6.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 3.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 16.7 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.2 | 2.3 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.2 | 1.5 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.2 | 1.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.2 | 3.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.2 | 9.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 0.5 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.2 | 26.9 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.2 | 2.5 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.2 | 10.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 2.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 1.0 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 3.0 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 50.1 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.1 | 23.3 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 5.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 3.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 3.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 10.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 9.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 6.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 3.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 2.0 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 1.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 5.2 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.1 | 0.3 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.6 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 29.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 3.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.6 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 0.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 2.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.3 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 19.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 4.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 5.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 5.4 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 5.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.5 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 1.6 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 18.1 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 4.7 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 1.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.6 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 195.3 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 2.1 | 59.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 1.7 | 55.7 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 1.4 | 111.6 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 1.3 | 24.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 1.2 | 40.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.8 | 28.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.8 | 18.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.7 | 6.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.7 | 22.0 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.6 | 25.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.5 | 5.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.5 | 29.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.4 | 171.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.3 | 19.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.3 | 0.6 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.3 | 7.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 9.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.2 | 2.1 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.2 | 4.8 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.2 | 3.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 3.3 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 9.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 8.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 5.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 10.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 2.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 5.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 2.1 | ST ADRENERGIC | Adrenergic Pathway |
| 0.1 | 3.2 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 2.7 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 4.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 2.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 4.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.3 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 3.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 2.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.6 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.4 | 8.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 3.6 | 239.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 3.4 | 73.9 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 2.7 | 144.1 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 2.0 | 18.3 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 1.4 | 13.7 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 1.3 | 7.5 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 1.2 | 25.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.9 | 18.0 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.8 | 26.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.8 | 18.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.7 | 49.9 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.5 | 8.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.5 | 11.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.5 | 13.0 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.4 | 9.3 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.4 | 17.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.4 | 12.7 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.3 | 7.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.3 | 27.2 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.3 | 7.5 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.2 | 7.2 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.2 | 10.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 1.6 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.2 | 9.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 11.4 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.2 | 3.6 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 6.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 4.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 2.5 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.2 | 3.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 6.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 3.9 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 4.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 13.9 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 11.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 9.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 7.3 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.1 | 3.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.7 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 6.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 2.4 | REACTOME PI METABOLISM | Genes involved in PI Metabolism |
| 0.1 | 2.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 3.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 0.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 5.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 2.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.1 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 2.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |