Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
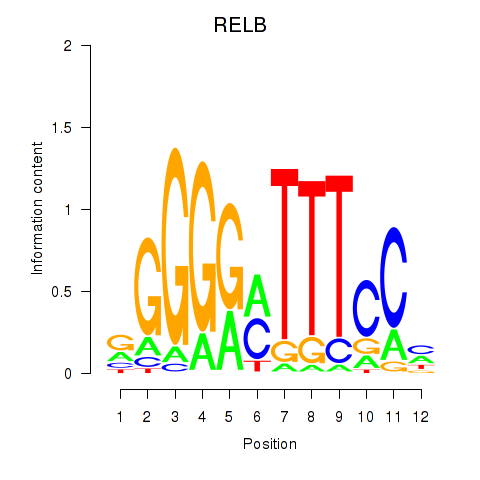
Results for RELB
Z-value: 1.19
Transcription factors associated with RELB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RELB
|
ENSG00000104856.9 | RELB proto-oncogene, NF-kB subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RELB | hg19_v2_chr19_+_45504688_45504782 | 0.81 | 1.1e-06 | Click! |
Activity profile of RELB motif
Sorted Z-values of RELB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_29527702 | 9.18 |
ENST00000377050.4
|
UBD
|
ubiquitin D |
| chr1_-_173020056 | 6.61 |
ENST00000239468.2
ENST00000404377.3 |
TNFSF18
|
tumor necrosis factor (ligand) superfamily, member 18 |
| chr4_+_74702214 | 5.82 |
ENST00000226317.5
ENST00000515050.1 |
CXCL6
|
chemokine (C-X-C motif) ligand 6 |
| chr1_+_110453109 | 5.49 |
ENST00000525659.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr4_-_74864386 | 4.94 |
ENST00000296027.4
|
CXCL5
|
chemokine (C-X-C motif) ligand 5 |
| chr14_+_103589789 | 4.92 |
ENST00000558056.1
ENST00000560869.1 |
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr19_+_45504688 | 4.39 |
ENST00000221452.8
ENST00000540120.1 ENST00000505236.1 |
RELB
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr4_+_103423055 | 4.06 |
ENST00000505458.1
|
NFKB1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr1_-_8000872 | 3.95 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr8_+_90770008 | 3.85 |
ENST00000540020.1
|
RIPK2
|
receptor-interacting serine-threonine kinase 2 |
| chr5_-_61031495 | 3.76 |
ENST00000506550.1
ENST00000512882.2 |
CTD-2170G1.2
|
CTD-2170G1.2 |
| chr6_+_138188551 | 3.75 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr6_-_32821599 | 3.69 |
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr5_-_150460914 | 3.69 |
ENST00000389378.2
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr11_+_102188224 | 3.63 |
ENST00000263464.3
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr1_+_110453203 | 3.62 |
ENST00000357302.4
ENST00000344188.5 ENST00000329608.6 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr5_-_150460539 | 3.62 |
ENST00000520931.1
ENST00000520695.1 ENST00000521591.1 ENST00000518977.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr12_-_49259643 | 3.61 |
ENST00000309739.5
|
RND1
|
Rho family GTPase 1 |
| chr1_+_110453462 | 3.61 |
ENST00000488198.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr11_+_102188272 | 3.43 |
ENST00000532808.1
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr10_+_104154229 | 3.38 |
ENST00000428099.1
ENST00000369966.3 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr8_+_90769967 | 3.28 |
ENST00000220751.4
|
RIPK2
|
receptor-interacting serine-threonine kinase 2 |
| chr6_+_32821924 | 3.21 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr5_-_146833222 | 3.16 |
ENST00000534907.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr11_+_19798964 | 2.88 |
ENST00000527559.2
|
NAV2
|
neuron navigator 2 |
| chr9_-_32526184 | 2.86 |
ENST00000545044.1
ENST00000379868.1 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr17_+_40440481 | 2.82 |
ENST00000590726.2
ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A
|
signal transducer and activator of transcription 5A |
| chr12_-_57504069 | 2.67 |
ENST00000543873.2
ENST00000554663.1 ENST00000557635.1 |
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr9_-_136344197 | 2.66 |
ENST00000414172.1
ENST00000371897.4 |
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr11_+_19799327 | 2.64 |
ENST00000540292.1
|
NAV2
|
neuron navigator 2 |
| chr6_+_132891461 | 2.40 |
ENST00000275198.1
|
TAAR6
|
trace amine associated receptor 6 |
| chr21_+_34775698 | 2.27 |
ENST00000381995.1
|
IFNGR2
|
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr4_-_185395672 | 2.27 |
ENST00000393593.3
|
IRF2
|
interferon regulatory factor 2 |
| chr21_+_34775772 | 2.21 |
ENST00000405436.1
|
IFNGR2
|
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr2_-_163175133 | 2.09 |
ENST00000421365.2
ENST00000263642.2 |
IFIH1
|
interferon induced with helicase C domain 1 |
| chr18_-_71959159 | 2.01 |
ENST00000494131.2
ENST00000397914.4 ENST00000340533.4 |
CYB5A
|
cytochrome b5 type A (microsomal) |
| chr6_+_80341000 | 1.97 |
ENST00000369838.4
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2 |
| chr16_+_50730910 | 1.92 |
ENST00000300589.2
|
NOD2
|
nucleotide-binding oligomerization domain containing 2 |
| chr2_+_68694678 | 1.80 |
ENST00000303795.4
|
APLF
|
aprataxin and PNKP like factor |
| chr8_+_54793454 | 1.62 |
ENST00000276500.4
|
RGS20
|
regulator of G-protein signaling 20 |
| chr8_+_54793425 | 1.62 |
ENST00000522225.1
|
RGS20
|
regulator of G-protein signaling 20 |
| chr14_+_103243813 | 1.58 |
ENST00000560371.1
ENST00000347662.4 ENST00000392745.2 ENST00000539721.1 ENST00000560463.1 |
TRAF3
|
TNF receptor-associated factor 3 |
| chr22_-_19512893 | 1.52 |
ENST00000403084.1
ENST00000413119.2 |
CLDN5
|
claudin 5 |
| chr10_+_13142075 | 1.52 |
ENST00000378757.2
ENST00000430081.1 ENST00000378752.3 ENST00000378748.3 |
OPTN
|
optineurin |
| chr5_-_146833485 | 1.49 |
ENST00000398514.3
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr2_+_32853093 | 1.49 |
ENST00000448773.1
ENST00000317907.4 |
TTC27
|
tetratricopeptide repeat domain 27 |
| chr10_+_13142225 | 1.47 |
ENST00000378747.3
|
OPTN
|
optineurin |
| chr5_-_59189545 | 1.41 |
ENST00000340635.6
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr16_+_53738053 | 1.36 |
ENST00000394647.3
|
FTO
|
fat mass and obesity associated |
| chr21_+_34775181 | 1.34 |
ENST00000290219.6
|
IFNGR2
|
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr16_-_2059748 | 1.34 |
ENST00000562103.1
ENST00000431526.1 |
ZNF598
|
zinc finger protein 598 |
| chr21_-_16437126 | 1.33 |
ENST00000318948.4
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr2_-_204400113 | 1.28 |
ENST00000319170.5
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr2_+_208394455 | 1.28 |
ENST00000430624.1
|
CREB1
|
cAMP responsive element binding protein 1 |
| chr21_-_16437255 | 1.27 |
ENST00000400199.1
ENST00000400202.1 |
NRIP1
|
nuclear receptor interacting protein 1 |
| chr10_+_13141585 | 1.24 |
ENST00000378764.2
|
OPTN
|
optineurin |
| chr11_-_65430554 | 1.23 |
ENST00000308639.9
ENST00000406246.3 |
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr4_+_74735102 | 1.21 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr9_+_72658490 | 1.19 |
ENST00000377182.4
|
MAMDC2
|
MAM domain containing 2 |
| chr2_+_26568965 | 1.18 |
ENST00000260585.7
ENST00000447170.1 |
EPT1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) |
| chr3_+_182971018 | 1.16 |
ENST00000326505.3
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr1_-_160313025 | 1.15 |
ENST00000368069.3
ENST00000241704.7 |
COPA
|
coatomer protein complex, subunit alpha |
| chr4_-_74904398 | 1.11 |
ENST00000296026.4
|
CXCL3
|
chemokine (C-X-C motif) ligand 3 |
| chr9_-_32526299 | 1.10 |
ENST00000379882.1
ENST00000379883.2 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr1_-_12677714 | 1.09 |
ENST00000376223.2
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr6_-_11779840 | 1.09 |
ENST00000506810.1
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr1_+_183441500 | 1.08 |
ENST00000456731.2
|
SMG7
|
SMG7 nonsense mediated mRNA decay factor |
| chr11_-_75062829 | 1.06 |
ENST00000393505.4
|
ARRB1
|
arrestin, beta 1 |
| chr1_-_95392635 | 1.06 |
ENST00000538964.1
ENST00000394202.4 ENST00000370206.4 |
CNN3
|
calponin 3, acidic |
| chr19_-_17958832 | 1.05 |
ENST00000458235.1
|
JAK3
|
Janus kinase 3 |
| chr10_-_75173785 | 1.05 |
ENST00000535178.1
ENST00000372921.5 ENST00000372919.4 |
ANXA7
|
annexin A7 |
| chr17_-_7590745 | 1.04 |
ENST00000514944.1
ENST00000503591.1 ENST00000455263.2 ENST00000420246.2 ENST00000445888.2 ENST00000509690.1 ENST00000604348.1 ENST00000269305.4 |
TP53
|
tumor protein p53 |
| chr10_-_96122682 | 1.01 |
ENST00000371361.3
|
NOC3L
|
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr15_-_77712477 | 1.01 |
ENST00000560626.2
|
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr15_+_52311398 | 1.01 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr3_-_47823298 | 0.98 |
ENST00000254480.5
|
SMARCC1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
| chr11_-_75062730 | 0.95 |
ENST00000420843.2
ENST00000360025.3 |
ARRB1
|
arrestin, beta 1 |
| chr8_+_38644715 | 0.93 |
ENST00000317827.4
ENST00000379931.3 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr6_+_29910301 | 0.91 |
ENST00000376809.5
ENST00000376802.2 |
HLA-A
|
major histocompatibility complex, class I, A |
| chr10_+_12391481 | 0.91 |
ENST00000378847.3
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID |
| chr1_+_183441600 | 0.89 |
ENST00000367537.3
|
SMG7
|
SMG7 nonsense mediated mRNA decay factor |
| chr17_-_77005860 | 0.89 |
ENST00000591773.1
ENST00000588611.1 ENST00000586916.2 ENST00000592033.1 ENST00000588075.1 ENST00000302345.2 ENST00000591811.1 |
CANT1
|
calcium activated nucleotidase 1 |
| chr10_+_114710425 | 0.86 |
ENST00000352065.5
ENST00000369395.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr12_-_54982300 | 0.84 |
ENST00000547431.1
|
PPP1R1A
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A |
| chr14_+_96858433 | 0.81 |
ENST00000267584.4
|
AK7
|
adenylate kinase 7 |
| chrX_+_108779870 | 0.79 |
ENST00000372107.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr1_+_110453514 | 0.79 |
ENST00000369802.3
ENST00000420111.2 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr2_+_232646379 | 0.78 |
ENST00000410024.1
ENST00000409295.1 ENST00000409091.1 |
COPS7B
|
COP9 signalosome subunit 7B |
| chr1_-_247242048 | 0.78 |
ENST00000366503.2
|
ZNF670
|
zinc finger protein 670 |
| chr17_-_77005801 | 0.76 |
ENST00000392446.5
|
CANT1
|
calcium activated nucleotidase 1 |
| chr22_-_37640456 | 0.76 |
ENST00000405484.1
ENST00000441619.1 ENST00000406508.1 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr8_-_101962777 | 0.74 |
ENST00000395951.3
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr6_-_36355513 | 0.73 |
ENST00000340181.4
ENST00000373737.4 |
ETV7
|
ets variant 7 |
| chr20_-_18477862 | 0.73 |
ENST00000337227.4
|
RBBP9
|
retinoblastoma binding protein 9 |
| chr16_+_57392684 | 0.72 |
ENST00000219235.4
|
CCL22
|
chemokine (C-C motif) ligand 22 |
| chr15_-_101817616 | 0.72 |
ENST00000526049.1
ENST00000398226.3 ENST00000537379.1 |
VIMP
|
VCP-interacting membrane protein |
| chr10_-_73975657 | 0.71 |
ENST00000394919.1
ENST00000526751.1 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr10_+_77056134 | 0.70 |
ENST00000528121.1
ENST00000416398.1 |
ZNF503-AS1
|
ZNF503 antisense RNA 1 |
| chrX_+_108780347 | 0.70 |
ENST00000372103.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr19_+_39390320 | 0.67 |
ENST00000576510.1
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr4_-_52904425 | 0.67 |
ENST00000535450.1
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| chr3_+_140770183 | 0.67 |
ENST00000310546.2
|
SPSB4
|
splA/ryanodine receptor domain and SOCS box containing 4 |
| chr6_+_15401075 | 0.66 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr2_-_100721923 | 0.66 |
ENST00000356421.2
|
AFF3
|
AF4/FMR2 family, member 3 |
| chr11_+_74951948 | 0.65 |
ENST00000562197.2
|
TPBGL
|
trophoblast glycoprotein-like |
| chr10_-_73976025 | 0.65 |
ENST00000342444.4
ENST00000533958.1 ENST00000527593.1 ENST00000394915.3 ENST00000530461.1 ENST00000317168.6 ENST00000524829.1 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr11_-_3862206 | 0.65 |
ENST00000351018.4
|
RHOG
|
ras homolog family member G |
| chr7_+_86273218 | 0.64 |
ENST00000361669.2
|
GRM3
|
glutamate receptor, metabotropic 3 |
| chr4_+_4861385 | 0.64 |
ENST00000382723.4
|
MSX1
|
msh homeobox 1 |
| chr4_+_39046615 | 0.62 |
ENST00000261425.3
ENST00000508137.2 |
KLHL5
|
kelch-like family member 5 |
| chr15_+_85923797 | 0.61 |
ENST00000559362.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr4_-_79860506 | 0.61 |
ENST00000295462.3
ENST00000380645.4 ENST00000512733.1 |
PAQR3
|
progestin and adipoQ receptor family member III |
| chr16_-_15188106 | 0.61 |
ENST00000429751.2
ENST00000564131.1 ENST00000563559.1 ENST00000198767.6 |
RRN3
|
RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) |
| chr16_-_2059797 | 0.60 |
ENST00000563630.1
|
ZNF598
|
zinc finger protein 598 |
| chr19_+_39390587 | 0.60 |
ENST00000572515.1
ENST00000392079.3 ENST00000575359.1 ENST00000313582.5 |
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr6_-_35656685 | 0.58 |
ENST00000539068.1
ENST00000540787.1 |
FKBP5
|
FK506 binding protein 5 |
| chr14_-_106967788 | 0.58 |
ENST00000390622.2
|
IGHV1-46
|
immunoglobulin heavy variable 1-46 |
| chr17_-_40828969 | 0.58 |
ENST00000591022.1
ENST00000587627.1 ENST00000293349.6 |
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr3_+_184032313 | 0.57 |
ENST00000392537.2
ENST00000444134.1 ENST00000450424.1 ENST00000421110.1 ENST00000382330.3 ENST00000426123.1 ENST00000350481.5 ENST00000455679.1 ENST00000440448.1 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr12_-_127256858 | 0.57 |
ENST00000542248.1
ENST00000540684.1 |
LINC00944
|
long intergenic non-protein coding RNA 944 |
| chr7_-_22539771 | 0.57 |
ENST00000406890.2
ENST00000424363.1 |
STEAP1B
|
STEAP family member 1B |
| chrX_-_65253506 | 0.56 |
ENST00000427538.1
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr1_+_24117693 | 0.56 |
ENST00000374503.3
ENST00000374502.3 |
LYPLA2
|
lysophospholipase II |
| chr5_+_67584174 | 0.56 |
ENST00000320694.8
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr12_+_6309517 | 0.56 |
ENST00000382519.4
ENST00000009180.4 |
CD9
|
CD9 molecule |
| chr7_-_69062391 | 0.56 |
ENST00000436600.2
|
RP5-942I16.1
|
RP5-942I16.1 |
| chr14_-_35873856 | 0.55 |
ENST00000553342.1
ENST00000216797.5 ENST00000557140.1 |
NFKBIA
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| chr21_-_43373999 | 0.54 |
ENST00000380486.3
|
C2CD2
|
C2 calcium-dependent domain containing 2 |
| chr11_+_64052266 | 0.54 |
ENST00000539851.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr1_+_37940153 | 0.53 |
ENST00000373087.6
|
ZC3H12A
|
zinc finger CCCH-type containing 12A |
| chr1_+_6845384 | 0.53 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr3_+_1134260 | 0.52 |
ENST00000446702.2
ENST00000539053.1 ENST00000350110.2 |
CNTN6
|
contactin 6 |
| chr2_+_114163945 | 0.52 |
ENST00000453673.3
|
IGKV1OR2-108
|
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chr12_-_53074182 | 0.52 |
ENST00000252244.3
|
KRT1
|
keratin 1 |
| chr16_+_3014217 | 0.51 |
ENST00000572045.1
|
KREMEN2
|
kringle containing transmembrane protein 2 |
| chr4_+_140222609 | 0.49 |
ENST00000296543.5
ENST00000398947.1 |
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr4_-_74964904 | 0.49 |
ENST00000508487.2
|
CXCL2
|
chemokine (C-X-C motif) ligand 2 |
| chr12_+_6308881 | 0.49 |
ENST00000382518.1
ENST00000536586.1 |
CD9
|
CD9 molecule |
| chr11_+_64052692 | 0.49 |
ENST00000377702.4
|
GPR137
|
G protein-coupled receptor 137 |
| chr1_+_75594119 | 0.49 |
ENST00000294638.5
|
LHX8
|
LIM homeobox 8 |
| chr11_+_64052294 | 0.49 |
ENST00000536667.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr7_+_6048856 | 0.48 |
ENST00000223029.3
ENST00000400479.2 ENST00000395236.2 |
AIMP2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr6_+_36164487 | 0.48 |
ENST00000357641.6
|
BRPF3
|
bromodomain and PHD finger containing, 3 |
| chrX_+_15518923 | 0.48 |
ENST00000348343.6
|
BMX
|
BMX non-receptor tyrosine kinase |
| chr5_+_134181625 | 0.47 |
ENST00000394976.3
|
C5orf24
|
chromosome 5 open reading frame 24 |
| chr11_+_5423827 | 0.47 |
ENST00000332043.1
|
OR51J1
|
olfactory receptor, family 51, subfamily J, member 1 (gene/pseudogene) |
| chr20_-_2451395 | 0.46 |
ENST00000339610.6
ENST00000381342.2 ENST00000438552.2 |
SNRPB
|
small nuclear ribonucleoprotein polypeptides B and B1 |
| chr14_+_29234870 | 0.45 |
ENST00000382535.3
|
FOXG1
|
forkhead box G1 |
| chr17_-_7141490 | 0.45 |
ENST00000574236.1
ENST00000572789.1 |
PHF23
|
PHD finger protein 23 |
| chr11_+_64052454 | 0.44 |
ENST00000539833.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr3_+_184032419 | 0.44 |
ENST00000352767.3
ENST00000427141.2 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr1_-_205904950 | 0.43 |
ENST00000340781.4
|
SLC26A9
|
solute carrier family 26 (anion exchanger), member 9 |
| chr16_+_2563871 | 0.43 |
ENST00000330398.4
ENST00000568562.1 ENST00000569317.1 |
ATP6V0C
ATP6C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c Uncharacterized protein |
| chr17_+_40985407 | 0.42 |
ENST00000586114.1
ENST00000590720.1 ENST00000585805.1 ENST00000541124.1 ENST00000441946.2 ENST00000591152.1 ENST00000589469.1 ENST00000293362.3 ENST00000592169.1 |
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki) |
| chr8_+_55528627 | 0.42 |
ENST00000220676.1
|
RP1
|
retinitis pigmentosa 1 (autosomal dominant) |
| chr10_+_86088381 | 0.41 |
ENST00000224756.8
ENST00000372088.2 |
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr2_-_27718052 | 0.41 |
ENST00000264703.3
|
FNDC4
|
fibronectin type III domain containing 4 |
| chr12_-_56221330 | 0.39 |
ENST00000546837.1
|
RP11-762I7.5
|
Uncharacterized protein |
| chrX_+_103031421 | 0.39 |
ENST00000433491.1
ENST00000418604.1 ENST00000443502.1 |
PLP1
|
proteolipid protein 1 |
| chr8_+_101170563 | 0.39 |
ENST00000520508.1
ENST00000388798.2 |
SPAG1
|
sperm associated antigen 1 |
| chr3_+_184032283 | 0.39 |
ENST00000346169.2
ENST00000414031.1 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr1_-_6321035 | 0.39 |
ENST00000377893.2
|
GPR153
|
G protein-coupled receptor 153 |
| chr3_+_148415571 | 0.38 |
ENST00000497524.1
ENST00000349243.3 ENST00000542281.1 ENST00000418473.2 ENST00000404754.2 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr2_+_64681103 | 0.38 |
ENST00000464281.1
|
LGALSL
|
lectin, galactoside-binding-like |
| chr12_-_125473600 | 0.38 |
ENST00000308736.2
|
DHX37
|
DEAH (Asp-Glu-Ala-His) box polypeptide 37 |
| chr3_-_107941230 | 0.38 |
ENST00000264538.3
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr3_+_42201653 | 0.38 |
ENST00000341421.3
ENST00000396175.1 |
TRAK1
|
trafficking protein, kinesin binding 1 |
| chr5_-_132073111 | 0.37 |
ENST00000403231.1
|
KIF3A
|
kinesin family member 3A |
| chr17_+_65373531 | 0.37 |
ENST00000580974.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr7_+_103969104 | 0.36 |
ENST00000424859.1
ENST00000535008.1 ENST00000401970.2 ENST00000543266.1 |
LHFPL3
|
lipoma HMGIC fusion partner-like 3 |
| chr4_-_76928641 | 0.36 |
ENST00000264888.5
|
CXCL9
|
chemokine (C-X-C motif) ligand 9 |
| chr20_+_9494987 | 0.36 |
ENST00000427562.2
ENST00000246070.2 |
LAMP5
|
lysosomal-associated membrane protein family, member 5 |
| chrX_+_108780062 | 0.36 |
ENST00000372106.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr6_+_42018614 | 0.36 |
ENST00000465926.1
ENST00000482432.1 |
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa |
| chr17_-_40540484 | 0.36 |
ENST00000588969.1
|
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr1_+_180165672 | 0.36 |
ENST00000443059.1
|
QSOX1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr1_-_75139397 | 0.35 |
ENST00000326665.5
|
C1orf173
|
chromosome 1 open reading frame 173 |
| chr2_+_159825143 | 0.35 |
ENST00000454300.1
ENST00000263635.6 |
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr6_-_24358264 | 0.34 |
ENST00000378454.3
|
DCDC2
|
doublecortin domain containing 2 |
| chr10_+_30722866 | 0.33 |
ENST00000263056.1
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr12_-_108991812 | 0.33 |
ENST00000392806.3
ENST00000547567.1 |
TMEM119
|
transmembrane protein 119 |
| chr12_-_31945172 | 0.33 |
ENST00000340398.3
|
H3F3C
|
H3 histone, family 3C |
| chr20_-_4795747 | 0.33 |
ENST00000379376.2
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2 |
| chr16_+_29819096 | 0.33 |
ENST00000568411.1
ENST00000563012.1 ENST00000562557.1 |
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr6_-_7389942 | 0.32 |
ENST00000296742.7
ENST00000502583.1 |
CAGE1
|
cancer antigen 1 |
| chr19_-_4065730 | 0.32 |
ENST00000601588.1
|
ZBTB7A
|
zinc finger and BTB domain containing 7A |
| chr3_-_194072019 | 0.32 |
ENST00000429275.1
ENST00000323830.3 |
CPN2
|
carboxypeptidase N, polypeptide 2 |
| chr17_-_40829026 | 0.31 |
ENST00000412503.1
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr1_+_94884023 | 0.31 |
ENST00000315713.5
|
ABCD3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr16_-_67978016 | 0.31 |
ENST00000264005.5
|
LCAT
|
lecithin-cholesterol acyltransferase |
| chr9_-_130693048 | 0.30 |
ENST00000388747.4
|
PIP5KL1
|
phosphatidylinositol-4-phosphate 5-kinase-like 1 |
| chr10_+_89419370 | 0.30 |
ENST00000361175.4
ENST00000456849.1 |
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr22_+_22786288 | 0.30 |
ENST00000390301.2
|
IGLV1-36
|
immunoglobulin lambda variable 1-36 |
| chrX_+_16737718 | 0.29 |
ENST00000380155.3
|
SYAP1
|
synapse associated protein 1 |
| chr1_-_202129704 | 0.28 |
ENST00000476061.1
ENST00000544762.1 ENST00000467283.1 ENST00000464870.1 ENST00000435759.2 ENST00000486116.1 ENST00000543735.1 ENST00000308986.5 ENST00000477625.1 |
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr3_-_155572164 | 0.28 |
ENST00000392845.3
ENST00000359479.3 |
SLC33A1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chrX_-_131352152 | 0.28 |
ENST00000342983.2
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr4_+_185395947 | 0.27 |
ENST00000605834.1
|
RP11-326I11.3
|
RP11-326I11.3 |
| chr18_-_70535177 | 0.27 |
ENST00000327305.6
|
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr1_-_113249734 | 0.27 |
ENST00000484054.3
ENST00000369636.2 ENST00000369637.1 ENST00000285735.2 ENST00000369638.2 |
RHOC
|
ras homolog family member C |
Network of associatons between targets according to the STRING database.
First level regulatory network of RELB
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 13.5 | GO:1904141 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 1.9 | 1.9 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 1.9 | 11.4 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 1.8 | 9.2 | GO:0070842 | aggresome assembly(GO:0070842) |
| 1.8 | 7.3 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 1.7 | 6.6 | GO:2000329 | negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 1.5 | 6.1 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.9 | 3.7 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.8 | 3.4 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.8 | 5.8 | GO:2000630 | positive regulation of miRNA metabolic process(GO:2000630) |
| 0.8 | 7.1 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.7 | 2.8 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.7 | 5.5 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.7 | 2.7 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.7 | 3.9 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.5 | 1.0 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.5 | 14.9 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.5 | 1.9 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.5 | 4.2 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.5 | 1.4 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.3 | 1.0 | GO:0002730 | regulation of dendritic cell cytokine production(GO:0002730) |
| 0.3 | 0.8 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.3 | 4.9 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.3 | 1.6 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.2 | 3.6 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.2 | 1.8 | GO:0051106 | positive regulation of DNA ligation(GO:0051106) |
| 0.2 | 1.3 | GO:0034670 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.2 | 0.6 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.2 | 2.8 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.2 | 1.5 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.2 | 2.6 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.2 | 2.0 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.2 | 3.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 0.5 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.2 | 1.4 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.2 | 0.9 | GO:0044334 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.2 | 0.3 | GO:0090107 | regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.1 | 0.6 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 | 1.4 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 4.8 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 1.4 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 2.0 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.1 | 0.3 | GO:0043132 | coenzyme A transport(GO:0015880) FAD transport(GO:0015883) coenzyme A transmembrane transport(GO:0035349) FAD transmembrane transport(GO:0035350) NAD transport(GO:0043132) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.1 | 0.4 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 0.3 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 0.9 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 0.5 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 1.0 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.1 | 0.6 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.1 | 0.3 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.1 | 0.3 | GO:0030451 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) |
| 0.1 | 0.5 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 2.7 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.6 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 1.1 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.1 | 0.3 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 1.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.5 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.5 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.1 | 0.3 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.1 | 0.3 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.1 | 0.2 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.1 | 0.2 | GO:0045357 | interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.1 | 0.2 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.1 | 0.2 | GO:2000213 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.1 | 0.2 | GO:0016999 | antibiotic metabolic process(GO:0016999) cellular amide catabolic process(GO:0043605) |
| 0.1 | 1.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.2 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.1 | 0.7 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.7 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.2 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.5 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.1 | GO:0016115 | terpenoid catabolic process(GO:0016115) |
| 0.0 | 0.8 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.5 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.6 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.4 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.2 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.0 | 0.4 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.7 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 0.4 | GO:0046549 | photoreceptor cell outer segment organization(GO:0035845) retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 3.7 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.0 | 1.1 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.0 | 0.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.4 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.0 | 0.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.2 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.2 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 0.2 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.1 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.6 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:2000196 | positive regulation of female gonad development(GO:2000196) |
| 0.0 | 0.2 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.3 | GO:1903012 | positive regulation of bone development(GO:1903012) |
| 0.0 | 0.4 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.7 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 1.7 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 0.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:0060022 | hard palate development(GO:0060022) |
| 0.0 | 0.4 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 3.7 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.1 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) neutrophil mediated killing of bacterium(GO:0070944) |
| 0.0 | 0.6 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.6 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.2 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.5 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 1.2 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 0.2 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.4 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 2.3 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.2 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.3 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 1.5 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.4 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.1 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 0.9 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.2 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.3 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.4 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.6 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 13.5 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 1.0 | 12.0 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.5 | 3.7 | GO:0042825 | TAP complex(GO:0042825) |
| 0.4 | 3.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.2 | 5.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 1.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.2 | 0.9 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 0.6 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 4.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 8.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.5 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.1 | 1.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 1.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.5 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 0.9 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.0 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 1.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 1.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 3.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.5 | GO:0005683 | U7 snRNP(GO:0005683) histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 1.0 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 1.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 1.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 3.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.0 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 2.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.2 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.4 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.0 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.3 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 3.9 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.3 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.0 | 0.1 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 1.1 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.5 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 1.0 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.0 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 1.1 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 21.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.9 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 3.7 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 13.5 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 1.2 | 5.8 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.9 | 13.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.7 | 2.0 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.6 | 7.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.6 | 1.7 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.5 | 3.7 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.5 | 1.4 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.4 | 9.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.4 | 2.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.4 | 4.7 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.4 | 1.2 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.3 | 14.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.2 | 0.6 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.2 | 2.7 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.2 | 1.3 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.2 | 7.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 1.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 1.0 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.1 | 0.6 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 3.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.4 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.9 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 1.9 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.1 | 3.9 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 3.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.3 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.3 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) FAD transmembrane transporter activity(GO:0015230) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.1 | 1.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 1.6 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.3 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.3 | GO:0004877 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.1 | 1.8 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 0.5 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.1 | 2.2 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 0.3 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 0.3 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.1 | 1.4 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 1.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.4 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.6 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 0.9 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.2 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.1 | 0.9 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.2 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.1 | 0.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 1.0 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.2 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.1 | 0.2 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.1 | 2.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.5 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.5 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 2.0 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 6.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 3.2 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 0.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 1.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 1.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 2.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.2 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 2.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 5.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.7 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 4.2 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.3 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 2.5 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 1.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 5.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.9 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 13.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.4 | 18.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.3 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.3 | 12.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 5.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 4.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 7.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 3.0 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 2.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.8 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 3.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 1.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 2.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 14.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.8 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.8 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 2.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.6 | PID MTOR 4PATHWAY | mTOR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.8 | REACTOME P75NTR SIGNALS VIA NFKB | Genes involved in p75NTR signals via NF-kB |
| 0.4 | 7.6 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.3 | 4.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.3 | 12.7 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.2 | 4.7 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.2 | 14.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 4.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 0.2 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.2 | 3.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 3.8 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 4.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 2.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 2.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 1.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 4.2 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.1 | 1.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 2.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 2.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 3.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 1.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.7 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 3.2 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 2.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |