Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
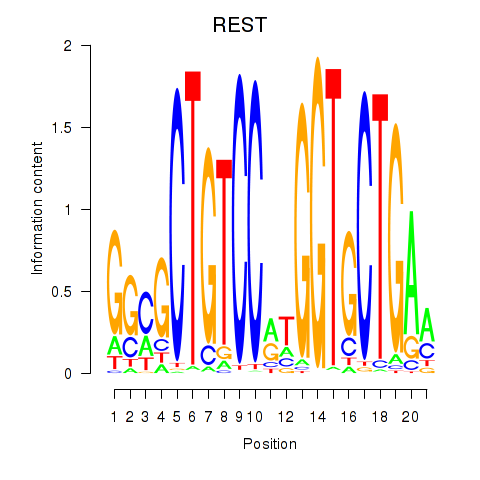
Results for REST
Z-value: 0.84
Transcription factors associated with REST
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
REST
|
ENSG00000084093.11 | RE1 silencing transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| REST | hg19_v2_chr4_+_57774042_57774114 | 0.26 | 2.0e-01 | Click! |
Activity profile of REST motif
Sorted Z-values of REST motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_93897272 | 2.22 |
ENST00000393151.2
|
UNC79
|
unc-79 homolog (C. elegans) |
| chr11_-_72492878 | 2.12 |
ENST00000535054.1
ENST00000545082.1 |
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr9_+_135457530 | 2.11 |
ENST00000263610.2
|
BARHL1
|
BarH-like homeobox 1 |
| chr14_+_93897199 | 1.57 |
ENST00000553484.1
|
UNC79
|
unc-79 homolog (C. elegans) |
| chr10_+_99609996 | 1.48 |
ENST00000370602.1
|
GOLGA7B
|
golgin A7 family, member B |
| chr4_+_8582287 | 1.46 |
ENST00000382487.4
|
GPR78
|
G protein-coupled receptor 78 |
| chr4_+_96012614 | 1.43 |
ENST00000264568.4
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr20_+_30555805 | 1.20 |
ENST00000562532.2
|
XKR7
|
XK, Kell blood group complex subunit-related family, member 7 |
| chr12_+_175930 | 1.09 |
ENST00000538872.1
ENST00000326261.4 |
IQSEC3
|
IQ motif and Sec7 domain 3 |
| chr18_+_57567180 | 1.05 |
ENST00000316660.6
ENST00000269518.9 |
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr19_-_40724246 | 1.03 |
ENST00000311308.6
|
TTC9B
|
tetratricopeptide repeat domain 9B |
| chr12_+_113229543 | 0.97 |
ENST00000447659.2
|
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr12_+_113229452 | 0.96 |
ENST00000389385.4
|
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr12_+_113229737 | 0.93 |
ENST00000551052.1
ENST00000415485.3 |
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr8_-_145559943 | 0.92 |
ENST00000332135.4
|
SCRT1
|
scratch family zinc finger 1 |
| chr9_-_139096955 | 0.86 |
ENST00000371748.5
|
LHX3
|
LIM homeobox 3 |
| chr5_+_33936491 | 0.79 |
ENST00000330120.3
|
RXFP3
|
relaxin/insulin-like family peptide receptor 3 |
| chr6_+_25652432 | 0.77 |
ENST00000377961.2
|
SCGN
|
secretagogin, EF-hand calcium binding protein |
| chr6_+_43603552 | 0.74 |
ENST00000372171.4
|
MAD2L1BP
|
MAD2L1 binding protein |
| chr1_+_212606219 | 0.73 |
ENST00000366988.3
|
NENF
|
neudesin neurotrophic factor |
| chr15_-_83378611 | 0.71 |
ENST00000542200.1
|
AP3B2
|
adaptor-related protein complex 3, beta 2 subunit |
| chr6_+_35773070 | 0.65 |
ENST00000373853.1
ENST00000360215.1 |
LHFPL5
|
lipoma HMGIC fusion partner-like 5 |
| chr10_-_79398250 | 0.63 |
ENST00000286627.5
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr22_+_23054174 | 0.63 |
ENST00000390308.2
|
IGLV3-21
|
immunoglobulin lambda variable 3-21 |
| chr6_+_154360616 | 0.62 |
ENST00000229768.5
ENST00000419506.2 ENST00000524163.1 ENST00000414028.2 ENST00000435918.2 ENST00000337049.4 |
OPRM1
|
opioid receptor, mu 1 |
| chr3_+_75955817 | 0.62 |
ENST00000487694.3
ENST00000602589.1 |
ROBO2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chr12_-_12837423 | 0.61 |
ENST00000540510.1
|
GPR19
|
G protein-coupled receptor 19 |
| chr20_-_44144249 | 0.60 |
ENST00000217428.6
|
SPINT3
|
serine peptidase inhibitor, Kunitz type, 3 |
| chr3_-_8686479 | 0.59 |
ENST00000544814.1
ENST00000427408.1 |
SSUH2
|
ssu-2 homolog (C. elegans) |
| chr6_+_154360553 | 0.58 |
ENST00000452687.2
|
OPRM1
|
opioid receptor, mu 1 |
| chrX_-_153237258 | 0.58 |
ENST00000310441.7
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr14_-_74892805 | 0.58 |
ENST00000331628.3
ENST00000554953.1 |
SYNDIG1L
|
synapse differentiation inducing 1-like |
| chr6_+_154360476 | 0.56 |
ENST00000428397.2
|
OPRM1
|
opioid receptor, mu 1 |
| chr4_-_175750364 | 0.53 |
ENST00000340217.5
ENST00000274093.3 |
GLRA3
|
glycine receptor, alpha 3 |
| chr18_-_40857493 | 0.52 |
ENST00000255224.3
|
SYT4
|
synaptotagmin IV |
| chr1_-_151689259 | 0.51 |
ENST00000420342.1
ENST00000290583.4 |
CELF3
|
CUGBP, Elav-like family member 3 |
| chr3_-_131753830 | 0.50 |
ENST00000429747.1
|
CPNE4
|
copine IV |
| chr5_+_71014990 | 0.50 |
ENST00000296777.4
|
CARTPT
|
CART prepropeptide |
| chrX_-_49056635 | 0.50 |
ENST00000472598.1
ENST00000538567.1 ENST00000479808.1 ENST00000263233.4 |
SYP
|
synaptophysin |
| chr8_-_57358432 | 0.48 |
ENST00000517415.1
ENST00000314922.3 |
PENK
|
proenkephalin |
| chr5_-_151304337 | 0.46 |
ENST00000455880.2
ENST00000545569.1 ENST00000274576.4 |
GLRA1
|
glycine receptor, alpha 1 |
| chr1_-_241520385 | 0.46 |
ENST00000366564.1
|
RGS7
|
regulator of G-protein signaling 7 |
| chr9_-_74061751 | 0.45 |
ENST00000357533.2
ENST00000423814.3 |
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr2_-_30143525 | 0.44 |
ENST00000431873.1
|
ALK
|
anaplastic lymphoma receptor tyrosine kinase |
| chr20_+_62327996 | 0.44 |
ENST00000369996.1
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy |
| chrX_-_153141302 | 0.44 |
ENST00000361699.4
ENST00000543994.1 ENST00000370057.3 ENST00000538883.1 ENST00000361981.3 |
L1CAM
|
L1 cell adhesion molecule |
| chr20_+_44657845 | 0.43 |
ENST00000243964.3
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5 |
| chr3_+_62304648 | 0.42 |
ENST00000462069.1
ENST00000232519.5 ENST00000465142.1 |
C3orf14
|
chromosome 3 open reading frame 14 |
| chr5_-_176037105 | 0.42 |
ENST00000303991.4
|
GPRIN1
|
G protein regulated inducer of neurite outgrowth 1 |
| chr10_+_60936921 | 0.41 |
ENST00000373878.3
|
PHYHIPL
|
phytanoyl-CoA 2-hydroxylase interacting protein-like |
| chr5_+_175298487 | 0.40 |
ENST00000393745.3
|
CPLX2
|
complexin 2 |
| chr9_+_140033862 | 0.37 |
ENST00000350902.5
ENST00000371550.4 ENST00000371546.4 ENST00000371555.4 ENST00000371553.3 ENST00000371559.4 ENST00000371560.3 |
GRIN1
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1 |
| chr20_-_62284766 | 0.37 |
ENST00000370053.1
|
STMN3
|
stathmin-like 3 |
| chr6_+_110299501 | 0.36 |
ENST00000414000.2
|
GPR6
|
G protein-coupled receptor 6 |
| chr3_+_62304712 | 0.35 |
ENST00000494481.1
|
C3orf14
|
chromosome 3 open reading frame 14 |
| chr2_-_51259641 | 0.34 |
ENST00000406316.2
ENST00000405581.1 |
NRXN1
|
neurexin 1 |
| chr11_-_125773085 | 0.33 |
ENST00000227474.3
ENST00000534158.1 ENST00000529801.1 |
PUS3
|
pseudouridylate synthase 3 |
| chr3_+_9745510 | 0.33 |
ENST00000383831.3
|
CPNE9
|
copine family member IX |
| chrX_-_47479246 | 0.31 |
ENST00000295987.7
ENST00000340666.4 |
SYN1
|
synapsin I |
| chr6_+_154360357 | 0.31 |
ENST00000330432.7
ENST00000360422.4 |
OPRM1
|
opioid receptor, mu 1 |
| chr5_-_135701164 | 0.30 |
ENST00000355180.3
ENST00000426057.2 ENST00000513104.1 |
TRPC7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chr10_-_44880491 | 0.29 |
ENST00000374426.2
ENST00000395795.4 ENST00000395794.2 ENST00000374429.2 ENST00000395793.3 ENST00000343575.6 |
CXCL12
|
chemokine (C-X-C motif) ligand 12 |
| chr12_-_99288536 | 0.29 |
ENST00000549797.1
ENST00000333732.7 ENST00000341752.7 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr13_-_111214015 | 0.29 |
ENST00000267328.3
|
RAB20
|
RAB20, member RAS oncogene family |
| chr14_-_94759595 | 0.28 |
ENST00000261994.4
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr2_-_51259528 | 0.28 |
ENST00000404971.1
|
NRXN1
|
neurexin 1 |
| chr17_-_3301704 | 0.28 |
ENST00000322608.2
|
OR1E1
|
olfactory receptor, family 1, subfamily E, member 1 |
| chr3_+_9745487 | 0.26 |
ENST00000383832.3
|
CPNE9
|
copine family member IX |
| chr14_-_94759408 | 0.26 |
ENST00000554723.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr4_+_158141899 | 0.25 |
ENST00000264426.9
ENST00000506284.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr12_-_57871853 | 0.24 |
ENST00000549602.1
ENST00000430041.2 |
ARHGAP9
|
Rho GTPase activating protein 9 |
| chr14_-_94759361 | 0.24 |
ENST00000393096.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr4_+_158141806 | 0.23 |
ENST00000393815.2
|
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr10_+_89419370 | 0.23 |
ENST00000361175.4
ENST00000456849.1 |
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr4_+_158141843 | 0.23 |
ENST00000509417.1
ENST00000296526.7 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr3_-_48700310 | 0.23 |
ENST00000164024.4
ENST00000544264.1 |
CELSR3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr20_+_36012051 | 0.22 |
ENST00000373567.2
|
SRC
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr1_-_102462565 | 0.22 |
ENST00000370103.4
|
OLFM3
|
olfactomedin 3 |
| chr9_-_104357277 | 0.21 |
ENST00000374806.1
|
PPP3R2
|
protein phosphatase 3, regulatory subunit B, beta |
| chr14_+_93389425 | 0.20 |
ENST00000216492.5
ENST00000334654.4 |
CHGA
|
chromogranin A (parathyroid secretory protein 1) |
| chr7_+_145813453 | 0.20 |
ENST00000361727.3
|
CNTNAP2
|
contactin associated protein-like 2 |
| chr20_-_60795296 | 0.20 |
ENST00000340177.5
|
HRH3
|
histamine receptor H3 |
| chr15_-_72612470 | 0.20 |
ENST00000287202.5
|
CELF6
|
CUGBP, Elav-like family member 6 |
| chr14_-_51561784 | 0.20 |
ENST00000360392.4
|
TRIM9
|
tripartite motif containing 9 |
| chr11_-_75141206 | 0.20 |
ENST00000376292.4
|
KLHL35
|
kelch-like family member 35 |
| chr11_-_92931098 | 0.20 |
ENST00000326402.4
|
SLC36A4
|
solute carrier family 36 (proton/amino acid symporter), member 4 |
| chr10_-_102790852 | 0.19 |
ENST00000470414.1
ENST00000370215.3 |
PDZD7
|
PDZ domain containing 7 |
| chr17_+_42385927 | 0.19 |
ENST00000426726.3
ENST00000590941.1 ENST00000225441.7 |
RUNDC3A
|
RUN domain containing 3A |
| chr11_+_22359562 | 0.18 |
ENST00000263160.3
|
SLC17A6
|
solute carrier family 17 (vesicular glutamate transporter), member 6 |
| chr2_-_47798044 | 0.18 |
ENST00000327876.4
|
KCNK12
|
potassium channel, subfamily K, member 12 |
| chr10_+_26505662 | 0.18 |
ENST00000428517.2
|
GAD2
|
glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) |
| chr5_+_63802109 | 0.17 |
ENST00000334025.2
|
RGS7BP
|
regulator of G-protein signaling 7 binding protein |
| chr17_-_7218403 | 0.17 |
ENST00000570780.1
|
GPS2
|
G protein pathway suppressor 2 |
| chr1_+_154540246 | 0.17 |
ENST00000368476.3
|
CHRNB2
|
cholinergic receptor, nicotinic, beta 2 (neuronal) |
| chr2_+_131513864 | 0.17 |
ENST00000458606.1
ENST00000423981.1 |
AMER3
|
APC membrane recruitment protein 3 |
| chr17_-_27332931 | 0.16 |
ENST00000442608.3
ENST00000335960.6 |
SEZ6
|
seizure related 6 homolog (mouse) |
| chr19_-_55691472 | 0.16 |
ENST00000537500.1
|
SYT5
|
synaptotagmin V |
| chr2_-_51259229 | 0.16 |
ENST00000405472.3
|
NRXN1
|
neurexin 1 |
| chr9_-_123812542 | 0.15 |
ENST00000223642.1
|
C5
|
complement component 5 |
| chr2_-_51259292 | 0.14 |
ENST00000401669.2
|
NRXN1
|
neurexin 1 |
| chr14_+_96039882 | 0.14 |
ENST00000556346.1
ENST00000553785.1 |
RP11-1070N10.4
|
RP11-1070N10.4 |
| chr12_+_71833550 | 0.14 |
ENST00000266674.5
|
LGR5
|
leucine-rich repeat containing G protein-coupled receptor 5 |
| chr11_+_45907177 | 0.13 |
ENST00000241014.2
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr8_-_91095099 | 0.12 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr3_-_42743006 | 0.12 |
ENST00000310417.5
|
HHATL
|
hedgehog acyltransferase-like |
| chr16_+_5008290 | 0.12 |
ENST00000251170.7
|
SEC14L5
|
SEC14-like 5 (S. cerevisiae) |
| chr1_-_40105617 | 0.11 |
ENST00000372852.3
|
HEYL
|
hes-related family bHLH transcription factor with YRPW motif-like |
| chr1_+_171750776 | 0.10 |
ENST00000458517.1
ENST00000362019.3 ENST00000367737.5 ENST00000361735.3 |
METTL13
|
methyltransferase like 13 |
| chr2_+_226265364 | 0.10 |
ENST00000272907.6
|
NYAP2
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
| chr7_+_150748288 | 0.10 |
ENST00000490540.1
|
ASIC3
|
acid-sensing (proton-gated) ion channel 3 |
| chr22_+_22749343 | 0.08 |
ENST00000390298.2
|
IGLV7-43
|
immunoglobulin lambda variable 7-43 |
| chr19_-_55691614 | 0.07 |
ENST00000592470.1
ENST00000354308.3 |
SYT5
|
synaptotagmin V |
| chr11_+_637246 | 0.06 |
ENST00000176183.5
|
DRD4
|
dopamine receptor D4 |
| chr2_+_115199876 | 0.06 |
ENST00000436732.1
ENST00000410059.1 |
DPP10
|
dipeptidyl-peptidase 10 (non-functional) |
| chr15_-_27018884 | 0.05 |
ENST00000299267.4
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr10_-_79398127 | 0.03 |
ENST00000372443.1
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr17_-_27333163 | 0.02 |
ENST00000360295.9
|
SEZ6
|
seizure related 6 homolog (mouse) |
| chr10_+_26505179 | 0.02 |
ENST00000376261.3
|
GAD2
|
glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) |
| chr3_-_113897545 | 0.02 |
ENST00000467632.1
|
DRD3
|
dopamine receptor D3 |
| chr15_-_22473353 | 0.02 |
ENST00000557788.2
|
IGHV4OR15-8
|
immunoglobulin heavy variable 4/OR15-8 (non-functional) |
| chr1_-_153517473 | 0.01 |
ENST00000368715.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr12_-_24737089 | 0.00 |
ENST00000483544.1
|
LINC00477
|
long intergenic non-protein coding RNA 477 |
| chr19_-_51845378 | 0.00 |
ENST00000335624.4
|
VSIG10L
|
V-set and immunoglobulin domain containing 10 like |
Network of associatons between targets according to the STRING database.
First level regulatory network of REST
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 0.3 | 0.9 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.3 | 1.4 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.3 | 3.9 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.2 | 2.1 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.2 | 0.5 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.2 | 0.6 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.1 | 1.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.6 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.1 | 0.4 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 0.4 | GO:0043449 | olfactory learning(GO:0008355) cellular alkene metabolic process(GO:0043449) |
| 0.1 | 0.5 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.1 | 0.7 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.1 | 0.4 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.8 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 1.0 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 0.9 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 | 1.1 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.1 | 0.2 | GO:2000705 | dense core granule biogenesis(GO:0061110) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 0.2 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.1 | 0.7 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.1 | 0.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 0.8 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.2 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.1 | 0.3 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.5 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.5 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 1.1 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.2 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.0 | 2.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.0 | 0.1 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.0 | 0.3 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.0 | 0.2 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.6 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.2 | GO:0032099 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.0 | 0.5 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 0.7 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.2 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.0 | 0.3 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.6 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.9 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.1 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.4 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.4 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.8 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.0 | 0.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 1.1 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 0.5 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.2 | 0.5 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.1 | 0.7 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.1 | 1.5 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 1.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 1.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.5 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.1 | 0.2 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.1 | 0.7 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 1.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.6 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 3.5 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0042583 | chromaffin granule(GO:0042583) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0004979 | beta-endorphin receptor activity(GO:0004979) morphine receptor activity(GO:0038047) |
| 0.3 | 2.9 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 1.0 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.2 | 1.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 0.8 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.7 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.5 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.6 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.2 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.4 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.2 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.1 | 0.9 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 1.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.7 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.4 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.5 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.4 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.6 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 1.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.2 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.4 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 1.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 2.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 1.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 1.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 1.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.0 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |