Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
Results for RFX7_RFX4_RFX1
Z-value: 0.89
Transcription factors associated with RFX7_RFX4_RFX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
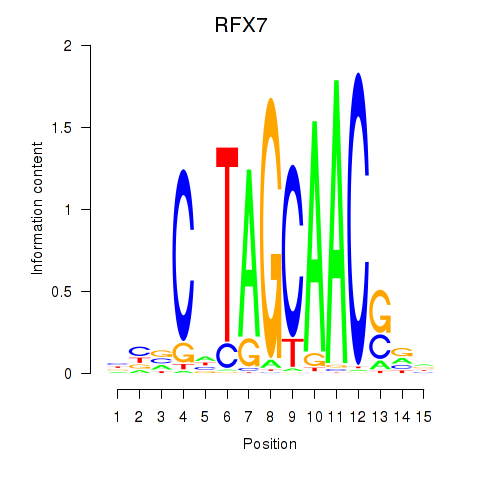
RFX7
|
ENSG00000181827.10 | regulatory factor X7 |
|
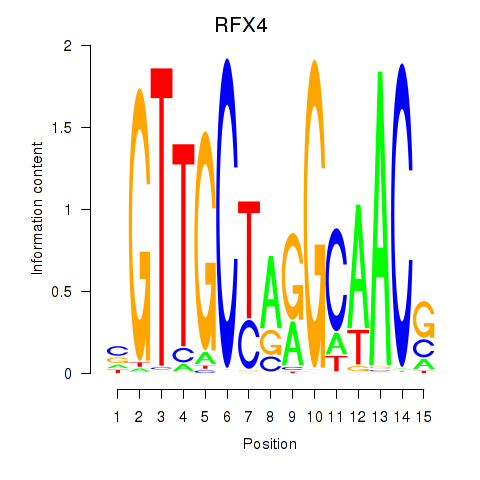
RFX4
|
ENSG00000111783.8 | regulatory factor X4 |
|
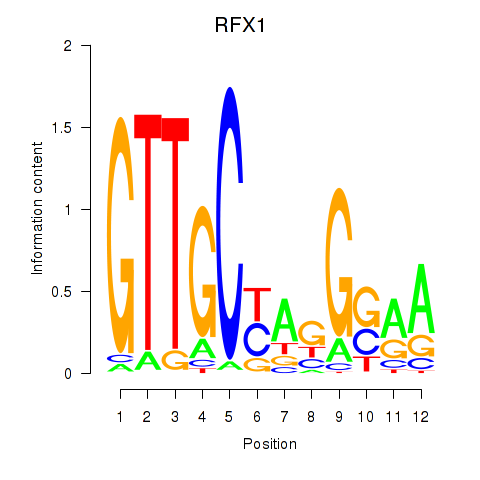
RFX1
|
ENSG00000132005.4 | regulatory factor X1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RFX7 | hg19_v2_chr15_-_56535464_56535521 | -0.24 | 2.5e-01 | Click! |
| RFX4 | hg19_v2_chr12_+_106976678_106976708, hg19_v2_chr12_+_107078474_107078533 | 0.09 | 6.7e-01 | Click! |
| RFX1 | hg19_v2_chr19_-_14117074_14117141 | 0.07 | 7.5e-01 | Click! |
Activity profile of RFX7_RFX4_RFX1 motif
Sorted Z-values of RFX7_RFX4_RFX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_50316517 | 1.79 |
ENST00000313777.4
ENST00000445575.2 |
FUZ
|
fuzzy planar cell polarity protein |
| chr19_-_50316489 | 1.63 |
ENST00000533418.1
|
FUZ
|
fuzzy planar cell polarity protein |
| chr19_-_50316423 | 1.60 |
ENST00000528094.1
ENST00000526575.1 |
FUZ
|
fuzzy planar cell polarity protein |
| chr16_+_57406368 | 1.56 |
ENST00000006053.6
ENST00000563383.1 |
CX3CL1
|
chemokine (C-X3-C motif) ligand 1 |
| chr10_-_25305011 | 1.49 |
ENST00000331161.4
ENST00000376363.1 |
ENKUR
|
enkurin, TRPC channel interacting protein |
| chr1_-_45308616 | 1.47 |
ENST00000447098.2
ENST00000372192.3 |
PTCH2
|
patched 2 |
| chr22_+_23487513 | 1.24 |
ENST00000263116.2
ENST00000341989.4 |
RAB36
|
RAB36, member RAS oncogene family |
| chr1_-_36916066 | 1.19 |
ENST00000315643.9
|
OSCP1
|
organic solute carrier partner 1 |
| chr19_+_16308711 | 1.18 |
ENST00000429941.2
ENST00000444449.2 ENST00000589822.1 |
AP1M1
|
adaptor-related protein complex 1, mu 1 subunit |
| chr19_-_51014345 | 0.98 |
ENST00000391815.3
ENST00000594350.1 ENST00000601423.1 |
JOSD2
|
Josephin domain containing 2 |
| chr20_-_30311703 | 0.97 |
ENST00000450273.1
ENST00000456404.1 ENST00000420488.1 ENST00000439267.1 |
BCL2L1
|
BCL2-like 1 |
| chr19_+_55888186 | 0.91 |
ENST00000291934.3
|
TMEM190
|
transmembrane protein 190 |
| chr20_-_30310797 | 0.88 |
ENST00000422920.1
|
BCL2L1
|
BCL2-like 1 |
| chr19_-_10679697 | 0.86 |
ENST00000335766.2
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr19_+_16308659 | 0.86 |
ENST00000590263.1
ENST00000590756.1 ENST00000541844.1 |
AP1M1
|
adaptor-related protein complex 1, mu 1 subunit |
| chr17_-_60885659 | 0.85 |
ENST00000311269.5
|
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr17_-_60885700 | 0.85 |
ENST00000583600.1
|
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr19_-_10679644 | 0.84 |
ENST00000393599.2
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr1_+_206808918 | 0.83 |
ENST00000367108.3
|
DYRK3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr1_+_206808868 | 0.80 |
ENST00000367109.2
|
DYRK3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr1_+_206809113 | 0.77 |
ENST00000441486.1
ENST00000367106.1 |
DYRK3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr11_+_61248583 | 0.68 |
ENST00000432063.2
ENST00000338608.2 |
PPP1R32
|
protein phosphatase 1, regulatory subunit 32 |
| chr11_-_113577052 | 0.65 |
ENST00000540540.1
ENST00000545579.1 ENST00000538955.1 ENST00000299882.5 |
TMPRSS5
|
transmembrane protease, serine 5 |
| chr1_-_21044489 | 0.63 |
ENST00000247986.2
|
KIF17
|
kinesin family member 17 |
| chr19_-_45953983 | 0.62 |
ENST00000592083.1
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr19_-_51014588 | 0.61 |
ENST00000598418.1
|
JOSD2
|
Josephin domain containing 2 |
| chr19_+_50321528 | 0.61 |
ENST00000312865.6
ENST00000595185.1 ENST00000538643.1 |
MED25
|
mediator complex subunit 25 |
| chrX_+_23925918 | 0.61 |
ENST00000379211.3
|
CXorf58
|
chromosome X open reading frame 58 |
| chr19_-_55791058 | 0.60 |
ENST00000587959.1
ENST00000585927.1 ENST00000587922.1 ENST00000585698.1 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr10_+_25305524 | 0.59 |
ENST00000524413.1
ENST00000376356.4 |
THNSL1
|
threonine synthase-like 1 (S. cerevisiae) |
| chr16_+_30773636 | 0.59 |
ENST00000402121.3
ENST00000565995.1 ENST00000563683.1 ENST00000357890.5 ENST00000565931.1 |
RNF40
|
ring finger protein 40, E3 ubiquitin protein ligase |
| chr2_-_170550877 | 0.56 |
ENST00000447353.1
|
CCDC173
|
coiled-coil domain containing 173 |
| chr19_-_41220957 | 0.53 |
ENST00000596357.1
ENST00000243583.6 ENST00000600080.1 ENST00000595254.1 ENST00000601967.1 |
ADCK4
|
aarF domain containing kinase 4 |
| chr16_+_57673207 | 0.53 |
ENST00000564783.1
ENST00000564729.1 ENST00000565976.1 ENST00000566508.1 ENST00000544297.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr19_-_51014460 | 0.52 |
ENST00000595669.1
|
JOSD2
|
Josephin domain containing 2 |
| chr20_+_814349 | 0.52 |
ENST00000381941.3
|
FAM110A
|
family with sequence similarity 110, member A |
| chr8_+_42010464 | 0.50 |
ENST00000518421.1
ENST00000174653.3 ENST00000396926.3 ENST00000521280.1 ENST00000522288.1 |
AP3M2
|
adaptor-related protein complex 3, mu 2 subunit |
| chr16_+_57673430 | 0.49 |
ENST00000540164.2
ENST00000568531.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr11_-_113577014 | 0.48 |
ENST00000544634.1
ENST00000539732.1 ENST00000538770.1 ENST00000536856.1 ENST00000544476.1 |
TMPRSS5
|
transmembrane protease, serine 5 |
| chr11_+_102188224 | 0.48 |
ENST00000263464.3
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chrX_-_23926004 | 0.48 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr1_+_161068179 | 0.47 |
ENST00000368011.4
ENST00000392192.2 |
KLHDC9
|
kelch domain containing 9 |
| chr4_-_177116772 | 0.46 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr12_-_76478686 | 0.46 |
ENST00000261182.8
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr19_-_40324767 | 0.44 |
ENST00000601972.1
ENST00000430012.2 ENST00000323039.5 ENST00000348817.3 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr5_+_68485433 | 0.44 |
ENST00000502689.1
|
CENPH
|
centromere protein H |
| chr19_-_3801789 | 0.44 |
ENST00000590849.1
ENST00000395045.2 |
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr9_-_138391692 | 0.44 |
ENST00000429260.2
|
C9orf116
|
chromosome 9 open reading frame 116 |
| chr1_+_36549676 | 0.43 |
ENST00000207457.3
|
TEKT2
|
tektin 2 (testicular) |
| chr15_+_67430339 | 0.43 |
ENST00000439724.3
|
SMAD3
|
SMAD family member 3 |
| chr20_-_10654639 | 0.42 |
ENST00000254958.5
|
JAG1
|
jagged 1 |
| chr11_+_69455855 | 0.42 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr7_-_62858860 | 0.41 |
ENST00000593633.1
|
AC073188.1
|
AC073188.1 |
| chr10_+_23216944 | 0.40 |
ENST00000298032.5
ENST00000409983.3 ENST00000409049.3 |
ARMC3
|
armadillo repeat containing 3 |
| chr22_+_38453207 | 0.40 |
ENST00000404072.3
ENST00000424694.1 |
PICK1
|
protein interacting with PRKCA 1 |
| chr11_+_113185533 | 0.40 |
ENST00000393020.1
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chrX_-_64254587 | 0.40 |
ENST00000337990.2
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr11_-_66103867 | 0.39 |
ENST00000424433.2
|
RIN1
|
Ras and Rab interactor 1 |
| chr22_+_38453378 | 0.39 |
ENST00000437453.1
ENST00000356976.3 |
PICK1
|
protein interacting with PRKCA 1 |
| chr10_+_23217006 | 0.38 |
ENST00000376528.4
ENST00000447081.1 |
ARMC3
|
armadillo repeat containing 3 |
| chr16_-_84538218 | 0.38 |
ENST00000562447.1
ENST00000565765.1 ENST00000535580.1 ENST00000343629.6 |
TLDC1
|
TBC/LysM-associated domain containing 1 |
| chr22_-_50970919 | 0.37 |
ENST00000329363.4
ENST00000437588.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr20_-_44540686 | 0.37 |
ENST00000477313.1
ENST00000542937.1 ENST00000372431.3 ENST00000354050.4 ENST00000420868.2 |
PLTP
|
phospholipid transfer protein |
| chr3_-_123339343 | 0.36 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr18_-_45935663 | 0.36 |
ENST00000589194.1
ENST00000591279.1 ENST00000590855.1 ENST00000587107.1 ENST00000588970.1 ENST00000586525.1 ENST00000592387.1 ENST00000590800.1 |
ZBTB7C
|
zinc finger and BTB domain containing 7C |
| chr14_+_96343100 | 0.36 |
ENST00000503525.2
|
LINC00617
|
long intergenic non-protein coding RNA 617 |
| chr12_+_113659234 | 0.36 |
ENST00000551096.1
ENST00000551099.1 ENST00000335509.6 ENST00000552897.1 ENST00000550785.1 ENST00000549279.1 |
TPCN1
|
two pore segment channel 1 |
| chr1_+_15736359 | 0.36 |
ENST00000375980.4
|
EFHD2
|
EF-hand domain family, member D2 |
| chr11_+_86085778 | 0.36 |
ENST00000354755.1
ENST00000278487.3 ENST00000531271.1 ENST00000445632.2 |
CCDC81
|
coiled-coil domain containing 81 |
| chr3_-_123339418 | 0.36 |
ENST00000583087.1
|
MYLK
|
myosin light chain kinase |
| chr1_-_118727781 | 0.35 |
ENST00000336338.5
|
SPAG17
|
sperm associated antigen 17 |
| chr1_-_36916011 | 0.34 |
ENST00000356637.5
ENST00000354267.3 ENST00000235532.5 |
OSCP1
|
organic solute carrier partner 1 |
| chr16_+_72127461 | 0.34 |
ENST00000268482.3
ENST00000566794.1 |
DHX38
|
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr17_-_80231300 | 0.33 |
ENST00000398519.5
ENST00000580446.1 |
CSNK1D
|
casein kinase 1, delta |
| chr14_+_105452094 | 0.32 |
ENST00000551606.1
ENST00000547315.1 |
C14orf79
|
chromosome 14 open reading frame 79 |
| chr5_-_149669192 | 0.32 |
ENST00000398376.3
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr11_-_85397167 | 0.32 |
ENST00000316398.3
|
CCDC89
|
coiled-coil domain containing 89 |
| chr16_+_72127670 | 0.32 |
ENST00000536867.1
|
DHX38
|
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr17_-_55911970 | 0.31 |
ENST00000581805.1
ENST00000580960.1 |
RP11-60A24.3
|
RP11-60A24.3 |
| chr21_-_44846999 | 0.31 |
ENST00000270162.6
|
SIK1
|
salt-inducible kinase 1 |
| chr17_-_15244894 | 0.31 |
ENST00000338696.2
ENST00000543896.1 ENST00000539245.1 ENST00000539316.1 ENST00000395930.1 |
TEKT3
|
tektin 3 |
| chr8_-_27115931 | 0.30 |
ENST00000523048.1
|
STMN4
|
stathmin-like 4 |
| chr7_-_102715263 | 0.30 |
ENST00000379305.3
|
FBXL13
|
F-box and leucine-rich repeat protein 13 |
| chr1_-_72748417 | 0.30 |
ENST00000357731.5
|
NEGR1
|
neuronal growth regulator 1 |
| chr12_+_111051832 | 0.29 |
ENST00000550703.2
ENST00000551590.1 |
TCTN1
|
tectonic family member 1 |
| chr2_+_219537015 | 0.29 |
ENST00000440309.1
ENST00000424080.1 |
STK36
|
serine/threonine kinase 36 |
| chr1_+_1447517 | 0.29 |
ENST00000378756.3
ENST00000378755.5 |
ATAD3A
|
ATPase family, AAA domain containing 3A |
| chr12_+_111051902 | 0.28 |
ENST00000397655.3
ENST00000471804.2 ENST00000377654.3 ENST00000397659.4 |
TCTN1
|
tectonic family member 1 |
| chr4_-_156297949 | 0.28 |
ENST00000515654.1
|
MAP9
|
microtubule-associated protein 9 |
| chr9_-_130635741 | 0.28 |
ENST00000223836.10
|
AK1
|
adenylate kinase 1 |
| chr12_-_47473707 | 0.28 |
ENST00000429635.1
|
AMIGO2
|
adhesion molecule with Ig-like domain 2 |
| chr7_+_86273218 | 0.28 |
ENST00000361669.2
|
GRM3
|
glutamate receptor, metabotropic 3 |
| chr22_-_23484246 | 0.28 |
ENST00000216036.4
|
RTDR1
|
rhabdoid tumor deletion region gene 1 |
| chr2_+_219536749 | 0.27 |
ENST00000295709.3
ENST00000392106.2 ENST00000392105.3 ENST00000455724.1 |
STK36
|
serine/threonine kinase 36 |
| chr6_+_32407619 | 0.27 |
ENST00000395388.2
ENST00000374982.5 |
HLA-DRA
|
major histocompatibility complex, class II, DR alpha |
| chr10_+_106113515 | 0.27 |
ENST00000369704.3
ENST00000312902.5 |
CCDC147
|
coiled-coil domain containing 147 |
| chr19_+_46850320 | 0.27 |
ENST00000391919.1
|
PPP5C
|
protein phosphatase 5, catalytic subunit |
| chr7_+_23637118 | 0.27 |
ENST00000448353.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr11_-_62521614 | 0.27 |
ENST00000527994.1
ENST00000394807.3 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chr4_+_96012614 | 0.27 |
ENST00000264568.4
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr15_+_74610894 | 0.26 |
ENST00000558821.1
ENST00000268082.4 |
CCDC33
|
coiled-coil domain containing 33 |
| chr22_-_30987837 | 0.26 |
ENST00000335214.6
|
PES1
|
pescadillo ribosomal biogenesis factor 1 |
| chr19_-_40324255 | 0.26 |
ENST00000593685.1
ENST00000600611.1 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr19_+_46850251 | 0.26 |
ENST00000012443.4
|
PPP5C
|
protein phosphatase 5, catalytic subunit |
| chr11_-_64885111 | 0.25 |
ENST00000528598.1
ENST00000310597.4 |
ZNHIT2
|
zinc finger, HIT-type containing 2 |
| chr1_-_74663825 | 0.25 |
ENST00000370911.3
ENST00000370909.2 ENST00000354431.4 |
LRRIQ3
|
leucine-rich repeats and IQ motif containing 3 |
| chr2_+_106682135 | 0.25 |
ENST00000437659.1
|
C2orf40
|
chromosome 2 open reading frame 40 |
| chr1_-_1051736 | 0.24 |
ENST00000448924.1
ENST00000294576.5 ENST00000437760.1 ENST00000462097.1 ENST00000475119.1 |
C1orf159
|
chromosome 1 open reading frame 159 |
| chr19_+_507299 | 0.24 |
ENST00000359315.5
|
TPGS1
|
tubulin polyglutamylase complex subunit 1 |
| chr19_+_19496728 | 0.24 |
ENST00000537887.1
ENST00000417582.2 |
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr6_+_116601265 | 0.24 |
ENST00000452085.3
|
DSE
|
dermatan sulfate epimerase |
| chr1_+_24120143 | 0.24 |
ENST00000374501.1
|
LYPLA2
|
lysophospholipase II |
| chr11_+_60681346 | 0.24 |
ENST00000227525.3
|
TMEM109
|
transmembrane protein 109 |
| chr2_-_190044480 | 0.23 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr2_+_219575543 | 0.23 |
ENST00000457313.1
ENST00000415717.1 ENST00000392102.1 |
TTLL4
|
tubulin tyrosine ligase-like family, member 4 |
| chr10_+_50887683 | 0.23 |
ENST00000374113.3
ENST00000374111.3 ENST00000374112.3 ENST00000535836.1 |
C10orf53
|
chromosome 10 open reading frame 53 |
| chr16_-_75590114 | 0.23 |
ENST00000568377.1
ENST00000565067.1 ENST00000258173.6 |
TMEM231
|
transmembrane protein 231 |
| chr1_-_36915880 | 0.23 |
ENST00000445843.3
|
OSCP1
|
organic solute carrier partner 1 |
| chr1_+_39456895 | 0.23 |
ENST00000432648.3
ENST00000446189.2 ENST00000372984.4 |
AKIRIN1
|
akirin 1 |
| chr9_+_138391805 | 0.23 |
ENST00000371785.1
|
MRPS2
|
mitochondrial ribosomal protein S2 |
| chr11_+_71791803 | 0.22 |
ENST00000539271.1
|
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr1_-_159893507 | 0.22 |
ENST00000368096.1
|
TAGLN2
|
transgelin 2 |
| chr2_+_27193480 | 0.22 |
ENST00000233121.2
ENST00000405074.3 |
MAPRE3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr19_-_55791563 | 0.22 |
ENST00000588971.1
ENST00000255631.5 ENST00000587551.1 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr17_-_56296580 | 0.22 |
ENST00000313863.6
ENST00000546108.1 ENST00000337050.7 ENST00000393119.2 |
MKS1
|
Meckel syndrome, type 1 |
| chr1_-_1051455 | 0.22 |
ENST00000379339.1
ENST00000480643.1 ENST00000434641.1 ENST00000421241.2 |
C1orf159
|
chromosome 1 open reading frame 159 |
| chr11_+_71791359 | 0.22 |
ENST00000419228.1
ENST00000435085.1 ENST00000307198.7 ENST00000538413.1 |
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr15_-_44969086 | 0.22 |
ENST00000434130.1
ENST00000560780.1 |
PATL2
|
protein associated with topoisomerase II homolog 2 (yeast) |
| chr6_-_32908765 | 0.22 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr14_-_39417410 | 0.21 |
ENST00000557019.1
ENST00000556116.1 ENST00000554732.1 |
LINC00639
|
long intergenic non-protein coding RNA 639 |
| chr20_+_814377 | 0.21 |
ENST00000304189.2
ENST00000381939.1 |
FAM110A
|
family with sequence similarity 110, member A |
| chr22_-_30987849 | 0.21 |
ENST00000402284.3
ENST00000354694.7 |
PES1
|
pescadillo ribosomal biogenesis factor 1 |
| chr22_-_43485381 | 0.21 |
ENST00000331018.7
ENST00000266254.7 ENST00000445824.1 |
TTLL1
|
tubulin tyrosine ligase-like family, member 1 |
| chr1_-_11865982 | 0.21 |
ENST00000418034.1
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr11_-_119991589 | 0.21 |
ENST00000526881.1
|
TRIM29
|
tripartite motif containing 29 |
| chr1_+_43637996 | 0.21 |
ENST00000528956.1
ENST00000529956.1 |
WDR65
|
WD repeat domain 65 |
| chr17_-_80231557 | 0.21 |
ENST00000392334.2
ENST00000314028.6 |
CSNK1D
|
casein kinase 1, delta |
| chr8_+_13424352 | 0.21 |
ENST00000297324.4
|
C8orf48
|
chromosome 8 open reading frame 48 |
| chr7_+_55177416 | 0.21 |
ENST00000450046.1
ENST00000454757.2 |
EGFR
|
epidermal growth factor receptor |
| chr17_-_45908875 | 0.21 |
ENST00000351111.2
ENST00000414011.1 |
MRPL10
|
mitochondrial ribosomal protein L10 |
| chr20_-_44718538 | 0.21 |
ENST00000290231.6
ENST00000372291.3 |
NCOA5
|
nuclear receptor coactivator 5 |
| chr2_+_54558004 | 0.20 |
ENST00000405749.1
ENST00000398634.2 ENST00000447328.1 |
C2orf73
|
chromosome 2 open reading frame 73 |
| chr19_+_572543 | 0.20 |
ENST00000333511.3
ENST00000573216.1 ENST00000353555.4 |
BSG
|
basigin (Ok blood group) |
| chr2_+_242089046 | 0.20 |
ENST00000407025.1
ENST00000272983.8 |
PPP1R7
|
protein phosphatase 1, regulatory subunit 7 |
| chr16_+_3550924 | 0.20 |
ENST00000576634.1
ENST00000574369.1 ENST00000341633.5 ENST00000417763.2 ENST00000571025.1 |
CLUAP1
|
clusterin associated protein 1 |
| chr15_+_71185148 | 0.20 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr12_+_49687425 | 0.20 |
ENST00000257860.4
|
PRPH
|
peripherin |
| chr6_+_52285131 | 0.20 |
ENST00000433625.2
|
EFHC1
|
EF-hand domain (C-terminal) containing 1 |
| chr8_+_10530155 | 0.20 |
ENST00000521818.1
|
C8orf74
|
chromosome 8 open reading frame 74 |
| chr20_+_62492566 | 0.20 |
ENST00000369916.3
|
ABHD16B
|
abhydrolase domain containing 16B |
| chr11_-_3078616 | 0.20 |
ENST00000401769.3
ENST00000278224.9 ENST00000397114.3 ENST00000380525.4 |
CARS
|
cysteinyl-tRNA synthetase |
| chr17_+_4736627 | 0.20 |
ENST00000355280.6
ENST00000347992.7 |
MINK1
|
misshapen-like kinase 1 |
| chr2_+_219187871 | 0.20 |
ENST00000258362.3
|
PNKD
|
paroxysmal nonkinesigenic dyskinesia |
| chr1_+_64669294 | 0.19 |
ENST00000371077.5
|
UBE2U
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr20_+_44420570 | 0.19 |
ENST00000372622.3
|
DNTTIP1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr4_+_123653807 | 0.19 |
ENST00000314218.3
ENST00000542236.1 |
BBS12
|
Bardet-Biedl syndrome 12 |
| chr4_+_15004165 | 0.19 |
ENST00000538197.1
ENST00000541112.1 ENST00000442003.2 |
CPEB2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr2_+_219575653 | 0.19 |
ENST00000442769.1
ENST00000424644.1 |
TTLL4
|
tubulin tyrosine ligase-like family, member 4 |
| chr17_+_73642486 | 0.19 |
ENST00000579469.1
|
SMIM6
|
small integral membrane protein 6 |
| chr19_-_55791540 | 0.19 |
ENST00000433386.2
|
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr17_+_73642315 | 0.19 |
ENST00000556126.2
|
SMIM6
|
small integral membrane protein 6 |
| chr3_+_111805182 | 0.19 |
ENST00000430855.1
ENST00000431717.2 ENST00000264848.5 |
C3orf52
|
chromosome 3 open reading frame 52 |
| chr1_+_150480551 | 0.18 |
ENST00000369049.4
ENST00000369047.4 |
ECM1
|
extracellular matrix protein 1 |
| chr22_+_18834324 | 0.18 |
ENST00000342005.4
|
AC008132.13
|
Uncharacterized protein |
| chr3_+_184056614 | 0.18 |
ENST00000453072.1
|
FAM131A
|
family with sequence similarity 131, member A |
| chr14_-_24658053 | 0.18 |
ENST00000354464.6
|
IPO4
|
importin 4 |
| chr15_+_71184931 | 0.18 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr2_-_217236750 | 0.18 |
ENST00000273067.4
|
MARCH4
|
membrane-associated ring finger (C3HC4) 4, E3 ubiquitin protein ligase |
| chr10_+_15001430 | 0.18 |
ENST00000407572.1
|
MEIG1
|
meiosis/spermiogenesis associated 1 |
| chr6_+_109761898 | 0.18 |
ENST00000258052.3
|
SMPD2
|
sphingomyelin phosphodiesterase 2, neutral membrane (neutral sphingomyelinase) |
| chr1_+_40627038 | 0.18 |
ENST00000372771.4
|
RLF
|
rearranged L-myc fusion |
| chr3_+_51705222 | 0.18 |
ENST00000457573.1
ENST00000341333.5 ENST00000412249.1 ENST00000425781.1 ENST00000415259.1 ENST00000395057.1 ENST00000416589.1 |
TEX264
|
testis expressed 264 |
| chr19_-_40971667 | 0.18 |
ENST00000263368.4
|
BLVRB
|
biliverdin reductase B (flavin reductase (NADPH)) |
| chr1_-_228603694 | 0.18 |
ENST00000366697.2
|
TRIM17
|
tripartite motif containing 17 |
| chr12_-_57472522 | 0.18 |
ENST00000379391.3
ENST00000300128.4 |
TMEM194A
|
transmembrane protein 194A |
| chr15_+_91473403 | 0.18 |
ENST00000394275.2
|
UNC45A
|
unc-45 homolog A (C. elegans) |
| chr12_+_119772502 | 0.18 |
ENST00000536742.1
ENST00000327554.2 |
CCDC60
|
coiled-coil domain containing 60 |
| chr22_+_31477296 | 0.17 |
ENST00000426927.1
ENST00000440425.1 ENST00000358743.1 ENST00000347557.2 ENST00000333137.7 |
SMTN
|
smoothelin |
| chr16_+_699319 | 0.17 |
ENST00000549091.1
ENST00000293879.4 |
WDR90
|
WD repeat domain 90 |
| chr20_+_44420617 | 0.17 |
ENST00000449078.1
ENST00000456939.1 |
DNTTIP1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr12_-_76478386 | 0.17 |
ENST00000535020.2
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr21_-_35883541 | 0.17 |
ENST00000399284.1
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1 |
| chr11_-_108422926 | 0.17 |
ENST00000428840.1
ENST00000526312.1 |
EXPH5
|
exophilin 5 |
| chr17_-_72709050 | 0.17 |
ENST00000583937.1
ENST00000301573.9 ENST00000326165.6 ENST00000469092.1 |
CD300LF
|
CD300 molecule-like family member f |
| chr12_-_76478446 | 0.17 |
ENST00000393263.3
ENST00000548044.1 ENST00000547704.1 ENST00000431879.3 ENST00000549596.1 ENST00000550934.1 ENST00000551600.1 ENST00000547479.1 ENST00000547773.1 ENST00000544816.1 ENST00000542344.1 ENST00000548273.1 |
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr9_-_34458531 | 0.17 |
ENST00000379089.1
ENST00000379087.1 ENST00000379084.1 ENST00000379081.1 ENST00000379080.1 ENST00000422409.1 ENST00000379078.1 ENST00000445726.1 ENST00000297620.4 |
FAM219A
|
family with sequence similarity 219, member A |
| chr7_+_149535455 | 0.17 |
ENST00000223210.4
ENST00000460379.1 |
ZNF862
|
zinc finger protein 862 |
| chr17_+_46985731 | 0.17 |
ENST00000360943.5
|
UBE2Z
|
ubiquitin-conjugating enzyme E2Z |
| chr17_-_8648105 | 0.17 |
ENST00000539522.2
ENST00000293845.3 |
CCDC42
|
coiled-coil domain containing 42 |
| chr5_+_68530697 | 0.17 |
ENST00000256443.3
ENST00000514676.1 |
CDK7
|
cyclin-dependent kinase 7 |
| chr7_+_99006550 | 0.17 |
ENST00000222969.5
|
BUD31
|
BUD31 homolog (S. cerevisiae) |
| chr12_+_48577366 | 0.17 |
ENST00000316554.3
|
C12orf68
|
chromosome 12 open reading frame 68 |
| chr19_-_11545920 | 0.17 |
ENST00000356392.4
ENST00000591179.1 |
CCDC151
|
coiled-coil domain containing 151 |
| chr3_-_107941209 | 0.17 |
ENST00000492106.1
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr19_+_2096868 | 0.17 |
ENST00000395296.1
ENST00000395301.3 |
IZUMO4
|
IZUMO family member 4 |
| chr22_-_50970506 | 0.17 |
ENST00000428989.2
ENST00000403326.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr19_-_10514184 | 0.17 |
ENST00000589629.1
ENST00000222005.2 |
CDC37
|
cell division cycle 37 |
| chr9_-_130477912 | 0.16 |
ENST00000543175.1
|
PTRH1
|
peptidyl-tRNA hydrolase 1 homolog (S. cerevisiae) |
| chr14_+_93357749 | 0.16 |
ENST00000557689.1
|
RP11-862G15.1
|
RP11-862G15.1 |
| chrX_+_139791917 | 0.16 |
ENST00000607004.1
ENST00000370535.3 |
LINC00632
|
long intergenic non-protein coding RNA 632 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RFX7_RFX4_RFX1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.0 | GO:0090299 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.5 | 1.6 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.5 | 2.4 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.2 | 1.8 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.2 | 0.6 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.2 | 1.5 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.2 | 0.6 | GO:0036034 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.1 | 0.6 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.1 | 1.7 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.1 | 0.4 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 0.6 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 | 0.3 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.1 | 0.8 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 0.6 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 | 0.4 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.1 | 0.3 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.1 | 0.6 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.1 | 0.3 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.1 | 0.1 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.1 | 2.0 | GO:0035646 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.1 | 0.9 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.5 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.4 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 0.6 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.1 | 0.7 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.2 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 | 0.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.1 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.1 | 0.2 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.3 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.1 | 0.5 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 0.2 | GO:0034125 | negative regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034125) regulation of interleukin-4-mediated signaling pathway(GO:1902214) |
| 0.1 | 0.6 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 0.2 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.3 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 0.1 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.1 | GO:0032888 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.1 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.2 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.0 | 0.1 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.5 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.0 | 0.1 | GO:1902868 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.0 | 0.2 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.0 | 0.4 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.3 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.1 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.4 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.1 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.0 | 0.1 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.0 | 0.1 | GO:0036114 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.2 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.2 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.1 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.0 | 0.5 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.3 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.0 | 0.4 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.5 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.5 | GO:0042424 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.2 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.0 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
| 0.0 | 0.1 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.1 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.1 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.0 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.4 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.0 | 0.3 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0035549 | positive regulation of interferon-beta secretion(GO:0035549) |
| 0.0 | 0.5 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.1 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.0 | 0.3 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.2 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.0 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.0 | 0.1 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 | 0.0 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 1.8 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.1 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.0 | GO:0090296 | regulation of mitochondrial DNA replication(GO:0090296) positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.0 | 0.2 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.6 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.2 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.4 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.7 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.2 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.0 | GO:0072615 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.0 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.0 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.1 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.2 | 0.9 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 0.6 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.1 | 0.4 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.1 | 1.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.8 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.3 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.1 | 0.6 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.3 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 0.2 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.1 | 0.5 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.7 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.1 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 2.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 1.3 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.7 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 2.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.0 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 0.6 | GO:0030990 | intraciliary transport particle(GO:0030990) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.1 | GO:0097165 | perichromatin fibrils(GO:0005726) nuclear stress granule(GO:0097165) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.4 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 1.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.2 | 1.8 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.3 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.1 | 0.9 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.7 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 0.4 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.4 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.3 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.1 | 1.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.2 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 0.3 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.1 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.1 | 0.5 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.3 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 3.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 1.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0033677 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) DNA/RNA helicase activity(GO:0033677) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.6 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 2.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.5 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.8 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.0 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.9 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.8 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.4 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 2.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.7 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.8 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 1.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.7 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 1.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.1 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |