Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
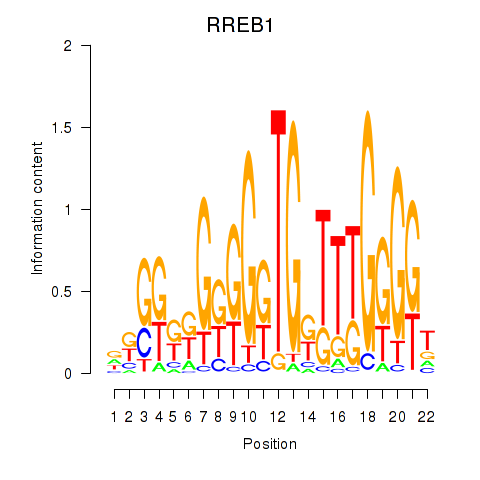
Results for RREB1
Z-value: 0.80
Transcription factors associated with RREB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RREB1
|
ENSG00000124782.15 | ras responsive element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RREB1 | hg19_v2_chr6_+_7107830_7107993, hg19_v2_chr6_+_7108210_7108264 | -0.22 | 2.9e-01 | Click! |
Activity profile of RREB1 motif
Sorted Z-values of RREB1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_61548374 | 4.04 |
ENST00000485903.2
ENST00000371185.2 ENST00000371184.2 |
NFIA
|
nuclear factor I/A |
| chr13_-_72441315 | 4.03 |
ENST00000305425.4
ENST00000313174.7 ENST00000354591.4 |
DACH1
|
dachshund homolog 1 (Drosophila) |
| chr1_+_61548225 | 3.30 |
ENST00000371187.3
|
NFIA
|
nuclear factor I/A |
| chr8_-_22550815 | 3.05 |
ENST00000317216.2
|
EGR3
|
early growth response 3 |
| chr12_+_96588279 | 2.81 |
ENST00000552142.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr2_+_48757278 | 1.84 |
ENST00000404752.1
ENST00000406226.1 |
STON1
|
stonin 1 |
| chr3_-_18466026 | 1.81 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr13_-_72440901 | 1.80 |
ENST00000359684.2
|
DACH1
|
dachshund homolog 1 (Drosophila) |
| chr1_+_164528866 | 1.77 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr9_-_14314066 | 1.74 |
ENST00000397575.3
|
NFIB
|
nuclear factor I/B |
| chr14_-_105635090 | 1.60 |
ENST00000331782.3
ENST00000347004.2 |
JAG2
|
jagged 2 |
| chr2_-_136873735 | 1.46 |
ENST00000409817.1
|
CXCR4
|
chemokine (C-X-C motif) receptor 4 |
| chr11_+_125034586 | 1.39 |
ENST00000298282.9
|
PKNOX2
|
PBX/knotted 1 homeobox 2 |
| chr9_-_14313893 | 1.37 |
ENST00000380921.3
ENST00000380959.3 |
NFIB
|
nuclear factor I/B |
| chr4_+_123747979 | 1.34 |
ENST00000608478.1
|
FGF2
|
fibroblast growth factor 2 (basic) |
| chr4_+_123747834 | 1.32 |
ENST00000264498.3
|
FGF2
|
fibroblast growth factor 2 (basic) |
| chr20_+_34700333 | 1.31 |
ENST00000441639.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr12_-_95044309 | 1.30 |
ENST00000261226.4
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr9_-_13279406 | 1.22 |
ENST00000546205.1
|
MPDZ
|
multiple PDZ domain protein |
| chr4_+_74735102 | 1.19 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr16_-_4588469 | 1.19 |
ENST00000588381.1
ENST00000563332.2 |
CDIP1
|
cell death-inducing p53 target 1 |
| chr20_+_62697564 | 1.15 |
ENST00000458442.1
|
TCEA2
|
transcription elongation factor A (SII), 2 |
| chr7_+_114562172 | 1.15 |
ENST00000393486.1
ENST00000257724.3 |
MDFIC
|
MyoD family inhibitor domain containing |
| chr20_-_36793663 | 1.14 |
ENST00000536701.1
ENST00000536724.1 |
TGM2
|
transglutaminase 2 |
| chr2_+_28618532 | 1.14 |
ENST00000545753.1
|
FOSL2
|
FOS-like antigen 2 |
| chr17_+_7788104 | 1.12 |
ENST00000380358.4
|
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr9_-_13279563 | 1.07 |
ENST00000541718.1
|
MPDZ
|
multiple PDZ domain protein |
| chr19_-_19051927 | 1.03 |
ENST00000600077.1
|
HOMER3
|
homer homolog 3 (Drosophila) |
| chr19_-_19051993 | 0.94 |
ENST00000594794.1
ENST00000355887.6 ENST00000392351.3 ENST00000596482.1 |
HOMER3
|
homer homolog 3 (Drosophila) |
| chr11_-_71791435 | 0.94 |
ENST00000351960.6
ENST00000541719.1 ENST00000535111.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr12_-_120687948 | 0.93 |
ENST00000458477.2
|
PXN
|
paxillin |
| chr11_-_71791518 | 0.92 |
ENST00000537217.1
ENST00000366394.3 ENST00000358965.6 ENST00000546131.1 ENST00000543937.1 ENST00000368959.5 ENST00000541641.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr11_-_89224488 | 0.92 |
ENST00000534731.1
ENST00000527626.1 |
NOX4
|
NADPH oxidase 4 |
| chr19_-_18995029 | 0.92 |
ENST00000596048.1
|
CERS1
|
ceramide synthase 1 |
| chr18_+_77160282 | 0.90 |
ENST00000318065.5
ENST00000545796.1 ENST00000592223.1 ENST00000329101.4 ENST00000586434.1 |
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr11_-_71791726 | 0.89 |
ENST00000393695.3
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr9_-_13279589 | 0.89 |
ENST00000319217.7
|
MPDZ
|
multiple PDZ domain protein |
| chr16_-_4588822 | 0.89 |
ENST00000564828.1
|
CDIP1
|
cell death-inducing p53 target 1 |
| chr17_-_39728303 | 0.85 |
ENST00000588431.1
ENST00000246662.4 |
KRT9
|
keratin 9 |
| chr16_-_4588762 | 0.84 |
ENST00000562334.1
ENST00000562579.1 ENST00000567695.1 ENST00000563507.1 |
CDIP1
|
cell death-inducing p53 target 1 |
| chr17_+_79373540 | 0.84 |
ENST00000307745.7
|
RP11-1055B8.7
|
BAH and coiled-coil domain-containing protein 1 |
| chr8_+_144816303 | 0.79 |
ENST00000533004.1
|
FAM83H-AS1
|
FAM83H antisense RNA 1 (head to head) |
| chr17_+_58499844 | 0.78 |
ENST00000269127.4
|
C17orf64
|
chromosome 17 open reading frame 64 |
| chr9_+_35906176 | 0.77 |
ENST00000354323.2
|
HRCT1
|
histidine rich carboxyl terminus 1 |
| chr10_-_126849588 | 0.77 |
ENST00000411419.2
|
CTBP2
|
C-terminal binding protein 2 |
| chr8_+_28748099 | 0.77 |
ENST00000519047.1
|
HMBOX1
|
homeobox containing 1 |
| chr13_-_37494365 | 0.75 |
ENST00000350148.5
|
SMAD9
|
SMAD family member 9 |
| chr13_-_37494391 | 0.73 |
ENST00000379826.4
|
SMAD9
|
SMAD family member 9 |
| chr7_-_132262060 | 0.73 |
ENST00000359827.3
|
PLXNA4
|
plexin A4 |
| chr16_-_49890016 | 0.73 |
ENST00000563137.2
|
ZNF423
|
zinc finger protein 423 |
| chr20_-_45984401 | 0.71 |
ENST00000311275.7
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr2_+_234104079 | 0.68 |
ENST00000417661.1
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr5_+_175299743 | 0.66 |
ENST00000502265.1
|
CPLX2
|
complexin 2 |
| chr3_-_39196049 | 0.64 |
ENST00000514182.1
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1 |
| chrX_+_118108571 | 0.63 |
ENST00000304778.7
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr9_+_96026230 | 0.62 |
ENST00000448251.1
|
WNK2
|
WNK lysine deficient protein kinase 2 |
| chr4_+_190992087 | 0.61 |
ENST00000553598.1
|
DUX4L7
|
double homeobox 4 like 7 |
| chr13_-_74708372 | 0.61 |
ENST00000377666.4
|
KLF12
|
Kruppel-like factor 12 |
| chr11_+_7506713 | 0.60 |
ENST00000329293.3
ENST00000534244.1 |
OLFML1
|
olfactomedin-like 1 |
| chr8_+_28748765 | 0.59 |
ENST00000355231.5
|
HMBOX1
|
homeobox containing 1 |
| chr17_-_7082861 | 0.59 |
ENST00000269299.3
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr14_+_57735614 | 0.58 |
ENST00000261558.3
|
AP5M1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr18_+_9334755 | 0.57 |
ENST00000262120.5
|
TWSG1
|
twisted gastrulation BMP signaling modulator 1 |
| chr19_-_3025614 | 0.57 |
ENST00000447365.2
|
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr6_+_37321748 | 0.56 |
ENST00000373479.4
ENST00000394443.4 |
RNF8
|
ring finger protein 8, E3 ubiquitin protein ligase |
| chr12_+_96588143 | 0.55 |
ENST00000228741.3
ENST00000547249.1 |
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr4_+_190995380 | 0.55 |
ENST00000555191.1
|
DUX4L6
|
double homeobox 4 like 6 |
| chr2_-_128432639 | 0.55 |
ENST00000545738.2
ENST00000355119.4 ENST00000409808.2 |
LIMS2
|
LIM and senescent cell antigen-like domains 2 |
| chr8_-_66754172 | 0.54 |
ENST00000401827.3
|
PDE7A
|
phosphodiesterase 7A |
| chr1_-_149889382 | 0.54 |
ENST00000369145.1
ENST00000369146.3 |
SV2A
|
synaptic vesicle glycoprotein 2A |
| chrX_+_17755563 | 0.54 |
ENST00000380045.3
ENST00000380041.3 ENST00000380043.3 ENST00000398080.1 |
SCML1
|
sex comb on midleg-like 1 (Drosophila) |
| chr4_+_190998973 | 0.52 |
ENST00000605696.1
ENST00000557283.2 |
DUX4L5
|
double homeobox 4 like 5 |
| chr3_-_47517302 | 0.52 |
ENST00000441517.2
ENST00000545718.1 |
SCAP
|
SREBF chaperone |
| chr4_+_190995680 | 0.52 |
ENST00000605381.1
ENST00000553722.2 |
DUX4L6
|
double homeobox 4 like 6 |
| chr6_-_13486369 | 0.52 |
ENST00000558378.1
|
AL583828.1
|
AL583828.1 |
| chr17_-_43502987 | 0.51 |
ENST00000376922.2
|
ARHGAP27
|
Rho GTPase activating protein 27 |
| chr1_-_21377383 | 0.51 |
ENST00000374935.3
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr4_+_190998673 | 0.51 |
ENST00000554668.1
|
DUX4L5
|
double homeobox 4 like 5 |
| chr4_+_190992387 | 0.51 |
ENST00000554906.2
ENST00000553820.2 |
DUX4L7
|
double homeobox 4 like 7 |
| chr4_+_191008860 | 0.51 |
ENST00000605191.1
ENST00000557656.2 |
DUX4L3
|
double homeobox 4 like 3 |
| chr4_+_191005567 | 0.51 |
ENST00000605758.1
ENST00000554285.2 |
DUX4
|
double homeobox 4 |
| chr4_+_191012160 | 0.51 |
ENST00000553702.1
ENST00000554376.1 |
DUX4L2
|
double homeobox 4 like 2 |
| chr4_+_191005267 | 0.51 |
ENST00000556625.1
|
DUX4
|
double homeobox 4 |
| chr4_+_191011860 | 0.51 |
ENST00000440426.1
|
DUX4L2
|
double homeobox 4 like 2 |
| chr4_+_191008560 | 0.51 |
ENST00000554690.1
|
DUX4L3
|
double homeobox 4 like 3 |
| chr19_-_42759300 | 0.51 |
ENST00000222329.4
|
ERF
|
Ets2 repressor factor |
| chr1_-_17307173 | 0.51 |
ENST00000438542.1
ENST00000375535.3 |
MFAP2
|
microfibrillar-associated protein 2 |
| chr19_-_3029011 | 0.50 |
ENST00000590536.1
ENST00000587137.1 ENST00000455444.2 ENST00000262953.6 |
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr1_-_94586651 | 0.49 |
ENST00000535735.1
ENST00000370225.3 |
ABCA4
|
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr19_-_48867291 | 0.48 |
ENST00000435956.3
|
TMEM143
|
transmembrane protein 143 |
| chr7_+_75932863 | 0.46 |
ENST00000429938.1
|
HSPB1
|
heat shock 27kDa protein 1 |
| chr19_+_34287751 | 0.46 |
ENST00000590771.1
ENST00000589786.1 ENST00000284006.6 ENST00000588881.1 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr17_+_6926339 | 0.45 |
ENST00000293805.5
|
BCL6B
|
B-cell CLL/lymphoma 6, member B |
| chr16_-_31439735 | 0.43 |
ENST00000287490.4
|
COX6A2
|
cytochrome c oxidase subunit VIa polypeptide 2 |
| chr20_+_33462888 | 0.42 |
ENST00000336325.4
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2 |
| chr17_-_39677971 | 0.42 |
ENST00000393976.2
|
KRT15
|
keratin 15 |
| chr16_-_28303360 | 0.42 |
ENST00000501520.1
|
RP11-57A19.2
|
RP11-57A19.2 |
| chr19_+_42381337 | 0.41 |
ENST00000597454.1
ENST00000444740.2 |
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr19_+_18544045 | 0.40 |
ENST00000599699.2
|
SSBP4
|
single stranded DNA binding protein 4 |
| chr16_-_75301886 | 0.40 |
ENST00000393422.2
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr7_-_752074 | 0.39 |
ENST00000360274.4
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chrX_+_151081351 | 0.39 |
ENST00000276344.2
|
MAGEA4
|
melanoma antigen family A, 4 |
| chr11_-_118436606 | 0.39 |
ENST00000530872.1
|
IFT46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr19_+_7828035 | 0.39 |
ENST00000327325.5
ENST00000394122.2 ENST00000248228.4 ENST00000334806.5 ENST00000359059.5 ENST00000357361.2 ENST00000596363.1 ENST00000595751.1 ENST00000596707.1 ENST00000597522.1 ENST00000595496.1 |
CLEC4M
|
C-type lectin domain family 4, member M |
| chr19_-_18653781 | 0.38 |
ENST00000596558.2
ENST00000453489.2 |
FKBP8
|
FK506 binding protein 8, 38kDa |
| chr7_+_95401851 | 0.37 |
ENST00000447467.2
|
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr12_-_8088871 | 0.37 |
ENST00000075120.7
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr12_-_6233828 | 0.36 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr17_+_35849937 | 0.36 |
ENST00000394389.4
|
DUSP14
|
dual specificity phosphatase 14 |
| chr12_-_7261772 | 0.36 |
ENST00000545280.1
ENST00000543933.1 ENST00000545337.1 ENST00000544702.1 ENST00000266542.4 |
C1RL
|
complement component 1, r subcomponent-like |
| chr16_+_58533951 | 0.35 |
ENST00000566192.1
ENST00000565088.1 ENST00000568640.1 ENST00000563978.1 ENST00000569923.1 ENST00000356752.4 ENST00000563799.1 ENST00000562999.1 ENST00000570248.1 ENST00000562731.1 ENST00000568424.1 |
NDRG4
|
NDRG family member 4 |
| chr5_+_150591678 | 0.35 |
ENST00000523466.1
|
GM2A
|
GM2 ganglioside activator |
| chrX_+_128674213 | 0.35 |
ENST00000371113.4
ENST00000357121.5 |
OCRL
|
oculocerebrorenal syndrome of Lowe |
| chr21_-_47648665 | 0.35 |
ENST00000450351.1
ENST00000522411.1 ENST00000356396.4 ENST00000397728.3 ENST00000457828.2 |
LSS
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) |
| chr13_-_76056250 | 0.35 |
ENST00000377636.3
ENST00000431480.2 ENST00000377625.2 ENST00000425511.1 |
TBC1D4
|
TBC1 domain family, member 4 |
| chr3_+_57094469 | 0.35 |
ENST00000334325.1
|
SPATA12
|
spermatogenesis associated 12 |
| chr16_+_68771128 | 0.34 |
ENST00000261769.5
ENST00000422392.2 |
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr19_+_55476620 | 0.33 |
ENST00000543010.1
ENST00000391721.4 ENST00000339757.7 |
NLRP2
|
NLR family, pyrin domain containing 2 |
| chr17_+_38474489 | 0.33 |
ENST00000394089.2
ENST00000425707.3 |
RARA
|
retinoic acid receptor, alpha |
| chr17_+_1733276 | 0.33 |
ENST00000254719.5
|
RPA1
|
replication protein A1, 70kDa |
| chr5_+_138629628 | 0.33 |
ENST00000508689.1
ENST00000514528.1 |
MATR3
|
matrin 3 |
| chr16_-_28503327 | 0.33 |
ENST00000535392.1
ENST00000395653.4 |
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr11_+_46402744 | 0.33 |
ENST00000533952.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr3_+_186435137 | 0.32 |
ENST00000447445.1
|
KNG1
|
kininogen 1 |
| chr7_+_91570165 | 0.32 |
ENST00000356239.3
ENST00000359028.2 ENST00000358100.2 |
AKAP9
|
A kinase (PRKA) anchor protein 9 |
| chr19_+_51358166 | 0.32 |
ENST00000601503.1
ENST00000326003.2 ENST00000597286.1 ENST00000597483.1 |
KLK3
|
kallikrein-related peptidase 3 |
| chr16_-_28503357 | 0.32 |
ENST00000333496.9
ENST00000561505.1 ENST00000567963.1 ENST00000354630.5 ENST00000355477.5 ENST00000357076.5 ENST00000565688.1 ENST00000359984.7 |
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr10_-_126849068 | 0.30 |
ENST00000494626.2
ENST00000337195.5 |
CTBP2
|
C-terminal binding protein 2 |
| chr5_-_35195338 | 0.30 |
ENST00000509839.1
|
PRLR
|
prolactin receptor |
| chrX_+_51636629 | 0.29 |
ENST00000375722.1
ENST00000326587.7 ENST00000375695.2 |
MAGED1
|
melanoma antigen family D, 1 |
| chr19_+_55477711 | 0.29 |
ENST00000448584.2
ENST00000537859.1 ENST00000585500.1 ENST00000427260.2 ENST00000538819.1 ENST00000263437.6 |
NLRP2
|
NLR family, pyrin domain containing 2 |
| chr16_-_28503080 | 0.29 |
ENST00000565316.1
ENST00000565778.1 ENST00000357857.9 ENST00000568558.1 ENST00000357806.7 |
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr7_-_71801980 | 0.28 |
ENST00000329008.5
|
CALN1
|
calneuron 1 |
| chr10_-_121356518 | 0.28 |
ENST00000369092.4
|
TIAL1
|
TIA1 cytotoxic granule-associated RNA binding protein-like 1 |
| chrX_-_148669116 | 0.28 |
ENST00000243314.5
|
MAGEA9B
|
melanoma antigen family A, 9B |
| chrX_+_102631844 | 0.27 |
ENST00000372634.1
ENST00000299872.7 |
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr4_-_148605265 | 0.27 |
ENST00000541232.1
ENST00000322396.6 |
PRMT10
|
protein arginine methyltransferase 10 (putative) |
| chr18_-_21242774 | 0.27 |
ENST00000322980.9
|
ANKRD29
|
ankyrin repeat domain 29 |
| chr13_+_60971427 | 0.27 |
ENST00000535286.1
ENST00000377881.2 |
TDRD3
|
tudor domain containing 3 |
| chr11_+_818902 | 0.26 |
ENST00000336615.4
|
PNPLA2
|
patatin-like phospholipase domain containing 2 |
| chrX_-_148571884 | 0.26 |
ENST00000537071.1
|
IDS
|
iduronate 2-sulfatase |
| chr9_-_116163400 | 0.26 |
ENST00000277315.5
ENST00000448137.1 ENST00000409155.3 |
ALAD
|
aminolevulinate dehydratase |
| chr1_+_24104869 | 0.25 |
ENST00000246151.4
|
PITHD1
|
PITH (C-terminal proteasome-interacting domain of thioredoxin-like) domain containing 1 |
| chr11_+_66406088 | 0.25 |
ENST00000310092.7
ENST00000396053.4 ENST00000408993.2 |
RBM4
|
RNA binding motif protein 4 |
| chr1_+_18958008 | 0.25 |
ENST00000420770.2
ENST00000400661.3 |
PAX7
|
paired box 7 |
| chr19_-_47975417 | 0.25 |
ENST00000236877.6
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr12_-_123756781 | 0.25 |
ENST00000544658.1
|
CDK2AP1
|
cyclin-dependent kinase 2 associated protein 1 |
| chr19_+_55999916 | 0.25 |
ENST00000587166.1
ENST00000389623.6 |
SSC5D
|
scavenger receptor cysteine rich domain containing (5 domains) |
| chr19_+_51358191 | 0.25 |
ENST00000593997.1
ENST00000595952.1 ENST00000360617.3 ENST00000598145.1 |
KLK3
|
kallikrein-related peptidase 3 |
| chr2_-_179343226 | 0.25 |
ENST00000434643.2
|
FKBP7
|
FK506 binding protein 7 |
| chr1_-_204380919 | 0.24 |
ENST00000367188.4
|
PPP1R15B
|
protein phosphatase 1, regulatory subunit 15B |
| chr6_+_37321823 | 0.23 |
ENST00000487950.1
ENST00000469731.1 |
RNF8
|
ring finger protein 8, E3 ubiquitin protein ligase |
| chr11_-_74109422 | 0.23 |
ENST00000298198.4
|
PGM2L1
|
phosphoglucomutase 2-like 1 |
| chr17_-_79817091 | 0.23 |
ENST00000570907.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr12_-_123756687 | 0.23 |
ENST00000261692.2
|
CDK2AP1
|
cyclin-dependent kinase 2 associated protein 1 |
| chr12_+_57916584 | 0.22 |
ENST00000546632.1
ENST00000549623.1 ENST00000431731.2 |
MBD6
|
methyl-CpG binding domain protein 6 |
| chrX_+_148863584 | 0.22 |
ENST00000439010.2
ENST00000298974.5 ENST00000522429.1 ENST00000519822.1 |
MAGEA9
|
melanoma antigen family A, 9 |
| chrX_+_151903207 | 0.22 |
ENST00000370287.3
|
CSAG1
|
chondrosarcoma associated gene 1 |
| chr15_-_23692381 | 0.21 |
ENST00000567107.1
ENST00000345070.5 ENST00000312015.5 |
GOLGA6L2
|
golgin A6 family-like 2 |
| chr7_+_94537542 | 0.21 |
ENST00000433881.1
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr7_+_39125365 | 0.21 |
ENST00000559001.1
ENST00000464276.2 |
POU6F2
|
POU class 6 homeobox 2 |
| chr5_+_139505520 | 0.21 |
ENST00000333305.3
|
IGIP
|
IgA-inducing protein |
| chr2_-_54087066 | 0.21 |
ENST00000394705.2
ENST00000352846.3 ENST00000406625.2 |
GPR75
GPR75-ASB3
ASB3
|
G protein-coupled receptor 75 GPR75-ASB3 readthrough Ankyrin repeat and SOCS box protein 3 |
| chr19_-_18654293 | 0.21 |
ENST00000597547.1
ENST00000222308.4 ENST00000544835.3 ENST00000610101.1 ENST00000597960.3 ENST00000608443.1 |
FKBP8
|
FK506 binding protein 8, 38kDa |
| chr14_+_100485712 | 0.21 |
ENST00000544450.2
|
EVL
|
Enah/Vasp-like |
| chr17_-_73761222 | 0.21 |
ENST00000437911.1
ENST00000225614.2 |
GALK1
|
galactokinase 1 |
| chr19_-_39226045 | 0.20 |
ENST00000597987.1
ENST00000595177.1 |
CAPN12
|
calpain 12 |
| chrX_+_153813407 | 0.20 |
ENST00000443287.2
ENST00000333128.3 |
CTAG1A
|
cancer/testis antigen 1A |
| chr10_-_95360983 | 0.20 |
ENST00000371464.3
|
RBP4
|
retinol binding protein 4, plasma |
| chr5_+_138629417 | 0.20 |
ENST00000510056.1
ENST00000511249.1 ENST00000503811.1 ENST00000511378.1 |
MATR3
|
matrin 3 |
| chr16_+_447209 | 0.20 |
ENST00000382940.4
ENST00000219479.2 |
NME4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr7_-_38398721 | 0.19 |
ENST00000390346.2
|
TRGV3
|
T cell receptor gamma variable 3 |
| chr11_+_61197572 | 0.19 |
ENST00000542074.1
ENST00000534878.1 ENST00000537782.1 ENST00000543265.1 |
SDHAF2
|
succinate dehydrogenase complex assembly factor 2 |
| chr17_-_7082668 | 0.18 |
ENST00000573083.1
ENST00000574388.1 |
ASGR1
|
asialoglycoprotein receptor 1 |
| chr12_+_57916466 | 0.18 |
ENST00000355673.3
|
MBD6
|
methyl-CpG binding domain protein 6 |
| chr1_-_226111929 | 0.18 |
ENST00000343818.6
ENST00000432920.2 |
PYCR2
RP4-559A3.7
|
pyrroline-5-carboxylate reductase family, member 2 Uncharacterized protein |
| chrX_+_151903253 | 0.18 |
ENST00000452779.2
ENST00000370291.2 |
CSAG1
|
chondrosarcoma associated gene 1 |
| chr17_+_58227287 | 0.17 |
ENST00000300900.4
ENST00000591725.1 |
CA4
|
carbonic anhydrase IV |
| chr11_-_61197480 | 0.17 |
ENST00000439958.3
ENST00000394888.4 |
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr12_+_49212514 | 0.17 |
ENST00000301050.2
ENST00000548279.1 ENST00000547230.1 |
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit |
| chr6_+_168227611 | 0.17 |
ENST00000344191.4
ENST00000351017.4 ENST00000392108.3 ENST00000366806.2 ENST00000392112.1 ENST00000400824.4 ENST00000447894.2 ENST00000400822.3 |
MLLT4
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 4 |
| chr2_+_219745020 | 0.16 |
ENST00000258411.3
|
WNT10A
|
wingless-type MMTV integration site family, member 10A |
| chr12_+_58003935 | 0.16 |
ENST00000333972.7
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chrX_-_77041685 | 0.16 |
ENST00000373344.5
ENST00000395603.3 |
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked |
| chr19_-_51192661 | 0.16 |
ENST00000391813.1
|
SHANK1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr12_+_16064258 | 0.16 |
ENST00000524480.1
ENST00000531803.1 ENST00000532964.1 |
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chr19_-_33555780 | 0.15 |
ENST00000254260.3
ENST00000400226.4 |
RHPN2
|
rhophilin, Rho GTPase binding protein 2 |
| chr3_+_41240986 | 0.15 |
ENST00000396185.3
|
CTNNB1
|
catenin (cadherin-associated protein), beta 1, 88kDa |
| chr2_+_85843252 | 0.15 |
ENST00000409025.1
ENST00000409470.1 ENST00000323701.6 ENST00000409766.3 |
USP39
|
ubiquitin specific peptidase 39 |
| chr14_-_71067360 | 0.15 |
ENST00000554963.1
ENST00000430055.2 ENST00000440435.2 ENST00000256379.5 |
MED6
|
mediator complex subunit 6 |
| chr7_+_95401877 | 0.14 |
ENST00000524053.1
ENST00000324972.6 ENST00000537881.1 ENST00000437599.1 ENST00000359388.4 ENST00000413338.1 |
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr11_+_61197508 | 0.14 |
ENST00000541135.1
ENST00000301761.2 |
RP11-286N22.8
SDHAF2
|
Uncharacterized protein succinate dehydrogenase complex assembly factor 2 |
| chrX_+_134166333 | 0.14 |
ENST00000257013.7
|
FAM127A
|
family with sequence similarity 127, member A |
| chr11_-_61197187 | 0.14 |
ENST00000449811.1
ENST00000413232.1 ENST00000340437.4 ENST00000539952.1 ENST00000544585.1 ENST00000450000.1 |
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr3_+_169684553 | 0.14 |
ENST00000337002.4
ENST00000480708.1 |
SEC62
|
SEC62 homolog (S. cerevisiae) |
| chr7_+_1022811 | 0.14 |
ENST00000308919.7
|
CYP2W1
|
cytochrome P450, family 2, subfamily W, polypeptide 1 |
| chr11_-_2323290 | 0.13 |
ENST00000381153.3
|
C11orf21
|
chromosome 11 open reading frame 21 |
| chr11_-_2323089 | 0.13 |
ENST00000456145.2
|
C11orf21
|
chromosome 11 open reading frame 21 |
| chr5_+_76145826 | 0.13 |
ENST00000513010.1
|
S100Z
|
S100 calcium binding protein Z |
| chr1_+_212782012 | 0.13 |
ENST00000341491.4
ENST00000366985.1 |
ATF3
|
activating transcription factor 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RREB1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:1902846 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.7 | 5.8 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.4 | 3.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.4 | 4.6 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.4 | 1.1 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.4 | 2.7 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.3 | 0.9 | GO:0072720 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.3 | 7.3 | GO:0072189 | ureter development(GO:0072189) |
| 0.2 | 0.7 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.2 | 0.9 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.2 | 0.7 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.2 | 1.5 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 1.0 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.5 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 | 1.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.7 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.1 | 0.5 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.1 | 0.3 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 0.2 | GO:0043385 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.1 | 0.4 | GO:0019541 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.1 | 0.4 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 0.5 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 0.3 | GO:0071284 | response to platinum ion(GO:0070541) cellular response to lead ion(GO:0071284) |
| 0.1 | 0.3 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 1.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.2 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.1 | 0.6 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.4 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.1 | 2.0 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 1.4 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 0.5 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.2 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.1 | 0.2 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.1 | 0.1 | GO:1901963 | regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) |
| 0.1 | 0.2 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.1 | 0.2 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.1 | 0.5 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.1 | 0.3 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.1 | 0.5 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.1 | 0.3 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 1.6 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.1 | 0.6 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 1.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.3 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 1.2 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.5 | GO:0002775 | antimicrobial peptide production(GO:0002775) antibacterial peptide production(GO:0002778) |
| 0.0 | 0.5 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.2 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.1 | GO:0050894 | determination of affect(GO:0050894) |
| 0.0 | 0.2 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.4 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 2.9 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 1.5 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.2 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.2 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 1.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.7 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.2 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.7 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.2 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.2 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.9 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 1.3 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.3 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.4 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.0 | 0.1 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.2 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.2 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.3 | GO:0036155 | positive regulation of triglyceride catabolic process(GO:0010898) acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.0 | 0.9 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 1.0 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 0.6 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.9 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.9 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.6 | GO:0050718 | positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.0 | 0.1 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.2 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 1.1 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 | 0.4 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of T-helper 2 cell differentiation(GO:0045629) negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.8 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.4 | 2.9 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.2 | 3.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 3.2 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 0.4 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.1 | 0.7 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 1.1 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 0.5 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 1.5 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 0.8 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.2 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 1.1 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.5 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 2.0 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.9 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 1.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.5 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.5 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 6.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.3 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.3 | 5.8 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.2 | 0.6 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.2 | 2.8 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.2 | 1.1 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 1.5 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 2.0 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.5 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.1 | 1.4 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 0.7 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 1.5 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 1.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.9 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.1 | 0.8 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 0.4 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 0.4 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.3 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.1 | 2.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.9 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.2 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.1 | 1.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 1.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 1.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.9 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.3 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 0.2 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.0 | 0.3 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.0 | 0.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.0 | 0.4 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.9 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.3 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.2 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.0 | 12.4 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 1.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.5 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.4 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.6 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 1.2 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.0 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.5 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 3.7 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.2 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 11.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 2.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 3.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 3.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 2.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.7 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.8 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.6 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 1.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.2 | 2.6 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 2.8 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.6 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 3.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.4 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.3 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.3 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 1.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.1 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.1 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |