Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
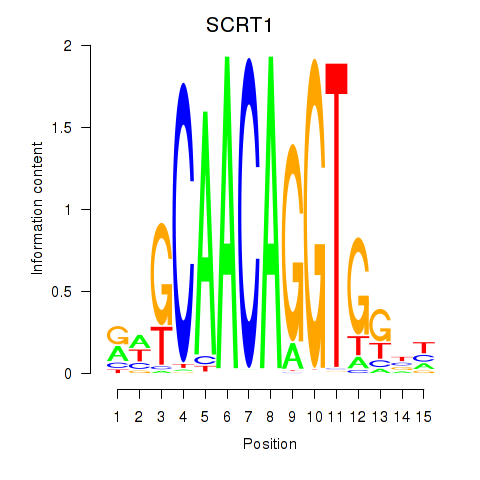
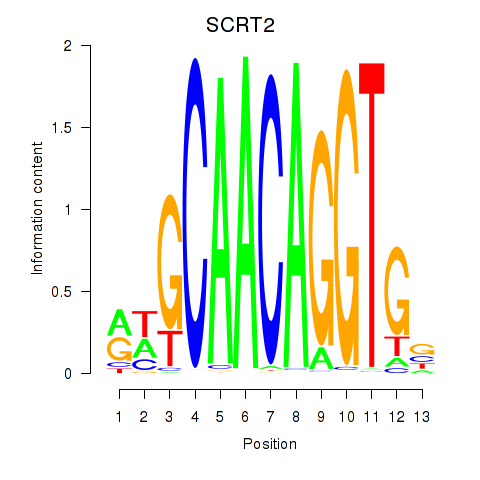
Results for SCRT1_SCRT2
Z-value: 0.57
Transcription factors associated with SCRT1_SCRT2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SCRT1
|
ENSG00000170616.9 | scratch family transcriptional repressor 1 |
|
SCRT2
|
ENSG00000215397.3 | scratch family transcriptional repressor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SCRT1 | hg19_v2_chr8_-_145559943_145559943 | -0.38 | 6.3e-02 | Click! |
Activity profile of SCRT1_SCRT2 motif
Sorted Z-values of SCRT1_SCRT2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_41540160 | 2.17 |
ENST00000503057.1
ENST00000511496.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr21_-_35987438 | 1.29 |
ENST00000313806.4
|
RCAN1
|
regulator of calcineurin 1 |
| chr10_+_35484793 | 1.05 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr4_-_90757364 | 1.00 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr7_+_114562172 | 0.93 |
ENST00000393486.1
ENST00000257724.3 |
MDFIC
|
MyoD family inhibitor domain containing |
| chr2_+_173600671 | 0.88 |
ENST00000409036.1
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr8_+_98881268 | 0.81 |
ENST00000254898.5
ENST00000524308.1 ENST00000522025.2 |
MATN2
|
matrilin 2 |
| chr6_-_10419871 | 0.77 |
ENST00000319516.4
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr16_+_67063262 | 0.75 |
ENST00000565389.1
|
CBFB
|
core-binding factor, beta subunit |
| chr4_-_10117949 | 0.72 |
ENST00000508079.1
|
WDR1
|
WD repeat domain 1 |
| chr10_+_35484053 | 0.71 |
ENST00000487763.1
ENST00000473940.1 ENST00000488328.1 ENST00000356917.5 |
CREM
|
cAMP responsive element modulator |
| chr17_+_78194205 | 0.70 |
ENST00000573809.1
ENST00000361193.3 ENST00000574967.1 ENST00000576126.1 ENST00000411502.3 ENST00000546047.2 |
SLC26A11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr20_-_50159198 | 0.64 |
ENST00000371564.3
ENST00000396009.3 ENST00000610033.1 |
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
| chr2_+_173600514 | 0.61 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr12_-_117318788 | 0.60 |
ENST00000550505.1
|
HRK
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr2_+_173600565 | 0.58 |
ENST00000397081.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr7_-_122526499 | 0.58 |
ENST00000412584.2
|
CADPS2
|
Ca++-dependent secretion activator 2 |
| chr11_+_61583721 | 0.50 |
ENST00000257261.6
|
FADS2
|
fatty acid desaturase 2 |
| chr2_+_101437487 | 0.50 |
ENST00000427413.1
ENST00000542504.1 |
NPAS2
|
neuronal PAS domain protein 2 |
| chr6_-_31697563 | 0.49 |
ENST00000375789.2
ENST00000416410.1 |
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr10_+_97515409 | 0.49 |
ENST00000371207.3
ENST00000543964.1 |
ENTPD1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr6_-_31697255 | 0.47 |
ENST00000436437.1
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr7_+_20370746 | 0.42 |
ENST00000222573.4
|
ITGB8
|
integrin, beta 8 |
| chr11_-_71159380 | 0.41 |
ENST00000525346.1
ENST00000531364.1 ENST00000529990.1 ENST00000527316.1 ENST00000407721.2 |
DHCR7
|
7-dehydrocholesterol reductase |
| chr8_+_120220561 | 0.41 |
ENST00000276681.6
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene) |
| chr10_+_6779326 | 0.39 |
ENST00000417112.1
|
RP11-554I8.2
|
RP11-554I8.2 |
| chr14_+_23776024 | 0.39 |
ENST00000553781.1
ENST00000556100.1 ENST00000557236.1 ENST00000557579.1 |
BCL2L2-PABPN1
BCL2L2
|
BCL2L2-PABPN1 readthrough BCL2-like 2 |
| chr1_+_55271736 | 0.38 |
ENST00000358193.3
ENST00000371273.3 |
C1orf177
|
chromosome 1 open reading frame 177 |
| chr6_+_90272027 | 0.38 |
ENST00000522441.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr14_+_23776167 | 0.38 |
ENST00000554635.1
ENST00000557008.1 |
BCL2L2
BCL2L2-PABPN1
|
BCL2-like 2 BCL2L2-PABPN1 readthrough |
| chr10_-_101945771 | 0.38 |
ENST00000370408.2
ENST00000407654.3 |
ERLIN1
|
ER lipid raft associated 1 |
| chr3_+_141144963 | 0.37 |
ENST00000510726.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chrX_+_55744228 | 0.36 |
ENST00000262850.7
|
RRAGB
|
Ras-related GTP binding B |
| chr2_-_24583583 | 0.35 |
ENST00000355123.4
|
ITSN2
|
intersectin 2 |
| chr16_+_67063855 | 0.35 |
ENST00000563939.2
|
CBFB
|
core-binding factor, beta subunit |
| chr8_-_133772794 | 0.35 |
ENST00000519187.1
ENST00000523829.1 ENST00000356838.3 ENST00000377901.4 ENST00000519304.1 |
TMEM71
|
transmembrane protein 71 |
| chr10_-_60027642 | 0.34 |
ENST00000373935.3
|
IPMK
|
inositol polyphosphate multikinase |
| chr13_+_32313658 | 0.34 |
ENST00000380314.1
ENST00000298386.2 |
RXFP2
|
relaxin/insulin-like family peptide receptor 2 |
| chr5_-_177423243 | 0.34 |
ENST00000308304.2
|
PROP1
|
PROP paired-like homeobox 1 |
| chr15_-_34628951 | 0.32 |
ENST00000397707.2
ENST00000560611.1 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr4_+_120133791 | 0.31 |
ENST00000274030.6
|
USP53
|
ubiquitin specific peptidase 53 |
| chr12_-_31478428 | 0.31 |
ENST00000543615.1
|
FAM60A
|
family with sequence similarity 60, member A |
| chr20_-_43977055 | 0.30 |
ENST00000372733.3
ENST00000537976.1 |
SDC4
|
syndecan 4 |
| chr19_-_18391708 | 0.30 |
ENST00000600972.1
|
JUND
|
jun D proto-oncogene |
| chr2_-_24583168 | 0.29 |
ENST00000361999.3
|
ITSN2
|
intersectin 2 |
| chr1_-_204135450 | 0.27 |
ENST00000272190.8
ENST00000367195.2 |
REN
|
renin |
| chr12_-_71003568 | 0.27 |
ENST00000547715.1
ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr3_+_49044798 | 0.26 |
ENST00000438660.1
ENST00000608424.1 ENST00000415265.2 |
WDR6
|
WD repeat domain 6 |
| chr5_+_125758813 | 0.25 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr11_+_64002292 | 0.25 |
ENST00000426086.2
|
VEGFB
|
vascular endothelial growth factor B |
| chr1_-_111217603 | 0.24 |
ENST00000369769.2
|
KCNA3
|
potassium voltage-gated channel, shaker-related subfamily, member 3 |
| chr11_+_44748361 | 0.24 |
ENST00000533202.1
ENST00000533080.1 ENST00000520358.2 ENST00000520999.2 |
TSPAN18
|
tetraspanin 18 |
| chr5_+_125758865 | 0.24 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr12_+_103981044 | 0.24 |
ENST00000388887.2
|
STAB2
|
stabilin 2 |
| chr20_-_45061695 | 0.24 |
ENST00000445496.2
|
ELMO2
|
engulfment and cell motility 2 |
| chrX_+_51928002 | 0.24 |
ENST00000375626.3
|
MAGED4
|
melanoma antigen family D, 4 |
| chrX_-_99986494 | 0.24 |
ENST00000372989.1
ENST00000455616.1 ENST00000454200.2 ENST00000276141.6 |
SYTL4
|
synaptotagmin-like 4 |
| chrX_-_51812268 | 0.24 |
ENST00000486010.1
ENST00000497164.1 ENST00000360134.6 ENST00000485287.1 ENST00000335504.5 ENST00000431659.1 |
MAGED4B
|
melanoma antigen family D, 4B |
| chrX_+_15518923 | 0.24 |
ENST00000348343.6
|
BMX
|
BMX non-receptor tyrosine kinase |
| chrX_+_51927919 | 0.23 |
ENST00000416960.1
|
MAGED4
|
melanoma antigen family D, 4 |
| chr2_+_62423242 | 0.23 |
ENST00000301998.4
|
B3GNT2
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 |
| chr22_+_44576770 | 0.23 |
ENST00000444313.3
ENST00000416291.1 |
PARVG
|
parvin, gamma |
| chr5_-_111091948 | 0.22 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr19_-_7990991 | 0.22 |
ENST00000318978.4
|
CTXN1
|
cortexin 1 |
| chrX_-_134232630 | 0.22 |
ENST00000535837.1
ENST00000433425.2 |
LINC00087
|
long intergenic non-protein coding RNA 87 |
| chr22_-_22090043 | 0.21 |
ENST00000403503.1
|
YPEL1
|
yippee-like 1 (Drosophila) |
| chrX_-_55057403 | 0.21 |
ENST00000396198.3
ENST00000335854.4 ENST00000455688.1 ENST00000330807.5 |
ALAS2
|
aminolevulinate, delta-, synthase 2 |
| chr1_+_222988406 | 0.20 |
ENST00000448808.1
ENST00000457636.1 ENST00000439440.1 |
RP11-452F19.3
|
RP11-452F19.3 |
| chr1_+_179851176 | 0.20 |
ENST00000528443.2
|
TOR1AIP1
|
torsin A interacting protein 1 |
| chr8_-_91657740 | 0.20 |
ENST00000422900.1
|
TMEM64
|
transmembrane protein 64 |
| chr3_+_151986709 | 0.20 |
ENST00000495875.2
ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr16_-_29757272 | 0.20 |
ENST00000329410.3
|
C16orf54
|
chromosome 16 open reading frame 54 |
| chr1_-_9129895 | 0.20 |
ENST00000473209.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chrX_-_70150939 | 0.19 |
ENST00000374299.3
ENST00000298085.4 |
SLC7A3
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3 |
| chr11_+_842808 | 0.19 |
ENST00000397397.2
ENST00000397411.2 ENST00000397396.1 |
TSPAN4
|
tetraspanin 4 |
| chr11_+_842928 | 0.19 |
ENST00000397408.1
|
TSPAN4
|
tetraspanin 4 |
| chr6_+_108977520 | 0.19 |
ENST00000540898.1
|
FOXO3
|
forkhead box O3 |
| chr18_+_52495426 | 0.18 |
ENST00000262094.5
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr3_-_179754556 | 0.17 |
ENST00000263962.8
|
PEX5L
|
peroxisomal biogenesis factor 5-like |
| chr3_-_72496035 | 0.17 |
ENST00000477973.2
|
RYBP
|
RING1 and YY1 binding protein |
| chrX_+_51636629 | 0.17 |
ENST00000375722.1
ENST00000326587.7 ENST00000375695.2 |
MAGED1
|
melanoma antigen family D, 1 |
| chr19_-_11373128 | 0.17 |
ENST00000294618.7
|
DOCK6
|
dedicator of cytokinesis 6 |
| chrX_+_118370211 | 0.17 |
ENST00000217971.7
|
PGRMC1
|
progesterone receptor membrane component 1 |
| chr8_+_82644669 | 0.16 |
ENST00000297265.4
|
CHMP4C
|
charged multivesicular body protein 4C |
| chr4_+_980785 | 0.16 |
ENST00000247933.4
ENST00000453894.1 |
IDUA
|
iduronidase, alpha-L- |
| chr1_+_16083154 | 0.16 |
ENST00000375771.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr6_-_27440460 | 0.16 |
ENST00000377419.1
|
ZNF184
|
zinc finger protein 184 |
| chr16_-_50402690 | 0.16 |
ENST00000394689.2
|
BRD7
|
bromodomain containing 7 |
| chr20_-_23402028 | 0.16 |
ENST00000398425.3
ENST00000432543.2 ENST00000377026.4 |
NAPB
|
N-ethylmaleimide-sensitive factor attachment protein, beta |
| chr22_+_44577237 | 0.16 |
ENST00000415224.1
ENST00000417767.1 |
PARVG
|
parvin, gamma |
| chr3_+_141144954 | 0.16 |
ENST00000441582.2
ENST00000321464.5 |
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr7_+_130126165 | 0.16 |
ENST00000427521.1
ENST00000416162.2 ENST00000378576.4 |
MEST
|
mesoderm specific transcript |
| chr1_+_222988464 | 0.16 |
ENST00000420335.1
|
RP11-452F19.3
|
RP11-452F19.3 |
| chr6_-_27440837 | 0.16 |
ENST00000211936.6
|
ZNF184
|
zinc finger protein 184 |
| chr17_+_53343577 | 0.15 |
ENST00000573945.1
|
HLF
|
hepatic leukemia factor |
| chr12_+_50794730 | 0.15 |
ENST00000523389.1
ENST00000518561.1 ENST00000347328.5 ENST00000550260.1 |
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr7_+_130126012 | 0.15 |
ENST00000341441.5
|
MEST
|
mesoderm specific transcript |
| chr19_+_35739782 | 0.15 |
ENST00000347609.4
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr19_+_35739897 | 0.15 |
ENST00000605618.1
ENST00000427250.1 ENST00000601623.1 |
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr9_-_126692386 | 0.15 |
ENST00000373624.2
ENST00000394219.3 ENST00000373620.3 ENST00000394215.2 ENST00000373618.1 |
DENND1A
|
DENN/MADD domain containing 1A |
| chr5_-_147211190 | 0.15 |
ENST00000510027.2
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1 |
| chr5_-_147211226 | 0.14 |
ENST00000296695.5
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1 |
| chr11_+_65779283 | 0.14 |
ENST00000312134.2
|
CST6
|
cystatin E/M |
| chr5_+_74011328 | 0.14 |
ENST00000513336.1
|
HEXB
|
hexosaminidase B (beta polypeptide) |
| chr19_+_35739597 | 0.13 |
ENST00000361790.3
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr19_+_12862604 | 0.13 |
ENST00000553030.1
|
BEST2
|
bestrophin 2 |
| chr20_+_30028322 | 0.13 |
ENST00000376309.3
|
DEFB123
|
defensin, beta 123 |
| chr17_-_40288449 | 0.13 |
ENST00000552162.1
ENST00000550504.1 |
RAB5C
|
RAB5C, member RAS oncogene family |
| chr19_+_35739631 | 0.13 |
ENST00000602003.1
ENST00000360798.3 ENST00000354900.3 |
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr19_+_35739280 | 0.13 |
ENST00000602122.1
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr12_+_16500037 | 0.13 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr16_+_103816 | 0.13 |
ENST00000383018.3
ENST00000417493.1 |
SNRNP25
|
small nuclear ribonucleoprotein 25kDa (U11/U12) |
| chr13_+_49551020 | 0.13 |
ENST00000541916.1
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr1_-_154928562 | 0.13 |
ENST00000368463.3
ENST00000539880.1 ENST00000542459.1 ENST00000368460.3 ENST00000368465.1 |
PBXIP1
|
pre-B-cell leukemia homeobox interacting protein 1 |
| chr1_-_94312706 | 0.13 |
ENST00000370244.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr15_-_30113676 | 0.13 |
ENST00000400011.2
|
TJP1
|
tight junction protein 1 |
| chr7_-_122526799 | 0.13 |
ENST00000334010.7
ENST00000313070.7 |
CADPS2
|
Ca++-dependent secretion activator 2 |
| chr17_-_2966901 | 0.12 |
ENST00000575751.1
|
OR1D5
|
olfactory receptor, family 1, subfamily D, member 5 |
| chr15_-_30114231 | 0.12 |
ENST00000356107.6
ENST00000545208.2 |
TJP1
|
tight junction protein 1 |
| chr14_+_23775971 | 0.12 |
ENST00000250405.5
|
BCL2L2
|
BCL2-like 2 |
| chr4_+_87856129 | 0.12 |
ENST00000395146.4
ENST00000507468.1 |
AFF1
|
AF4/FMR2 family, member 1 |
| chr16_+_2286726 | 0.11 |
ENST00000382437.4
ENST00000569184.1 |
DNASE1L2
|
deoxyribonuclease I-like 2 |
| chr4_-_140223670 | 0.11 |
ENST00000394228.1
ENST00000539387.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr12_-_67197760 | 0.11 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr10_-_47222824 | 0.11 |
ENST00000355232.3
|
AGAP10
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 10 |
| chr1_+_150245099 | 0.11 |
ENST00000369099.3
|
C1orf54
|
chromosome 1 open reading frame 54 |
| chr19_-_51220176 | 0.11 |
ENST00000359082.3
ENST00000293441.1 |
SHANK1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr2_-_61244308 | 0.11 |
ENST00000407787.1
ENST00000398658.2 |
PUS10
|
pseudouridylate synthase 10 |
| chr17_+_39261584 | 0.10 |
ENST00000391415.1
|
KRTAP4-9
|
keratin associated protein 4-9 |
| chrX_-_99987088 | 0.10 |
ENST00000372981.1
ENST00000263033.5 |
SYTL4
|
synaptotagmin-like 4 |
| chr5_+_72921983 | 0.10 |
ENST00000296794.6
ENST00000545377.1 ENST00000513042.2 ENST00000287898.5 ENST00000509848.1 |
ARHGEF28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr5_+_148651409 | 0.10 |
ENST00000296721.4
|
AFAP1L1
|
actin filament associated protein 1-like 1 |
| chrX_-_151143140 | 0.10 |
ENST00000393914.3
ENST00000370328.3 ENST00000370325.1 |
GABRE
|
gamma-aminobutyric acid (GABA) A receptor, epsilon |
| chr13_-_25746416 | 0.10 |
ENST00000515384.1
ENST00000357816.2 |
AMER2
|
APC membrane recruitment protein 2 |
| chr4_-_83483395 | 0.10 |
ENST00000515780.2
|
TMEM150C
|
transmembrane protein 150C |
| chr4_-_110223523 | 0.09 |
ENST00000399127.1
|
COL25A1
|
collagen, type XXV, alpha 1 |
| chr8_-_91657909 | 0.09 |
ENST00000418210.2
|
TMEM64
|
transmembrane protein 64 |
| chr1_-_224033596 | 0.09 |
ENST00000391878.2
ENST00000343537.7 |
TP53BP2
|
tumor protein p53 binding protein, 2 |
| chr1_+_20617362 | 0.09 |
ENST00000375079.2
ENST00000289815.8 ENST00000375083.4 ENST00000289825.4 |
VWA5B1
|
von Willebrand factor A domain containing 5B1 |
| chrX_+_13707235 | 0.09 |
ENST00000464506.1
|
RAB9A
|
RAB9A, member RAS oncogene family |
| chr16_-_1031259 | 0.09 |
ENST00000563837.1
ENST00000563863.1 ENST00000565069.1 ENST00000570014.1 |
RP11-161M6.2
LMF1
|
RP11-161M6.2 lipase maturation factor 1 |
| chr4_+_87856191 | 0.08 |
ENST00000503477.1
|
AFF1
|
AF4/FMR2 family, member 1 |
| chr3_-_49851313 | 0.08 |
ENST00000333486.3
|
UBA7
|
ubiquitin-like modifier activating enzyme 7 |
| chr20_+_57427765 | 0.08 |
ENST00000371100.4
|
GNAS
|
GNAS complex locus |
| chr14_-_71107921 | 0.08 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr17_-_34257731 | 0.08 |
ENST00000431884.2
ENST00000425909.3 ENST00000394528.3 ENST00000430160.2 |
RDM1
|
RAD52 motif 1 |
| chr22_-_22090064 | 0.08 |
ENST00000339468.3
|
YPEL1
|
yippee-like 1 (Drosophila) |
| chr17_-_79805146 | 0.07 |
ENST00000415593.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr22_-_30662828 | 0.07 |
ENST00000403463.1
ENST00000215781.2 |
OSM
|
oncostatin M |
| chr19_-_3062881 | 0.07 |
ENST00000586742.1
|
AES
|
amino-terminal enhancer of split |
| chr19_-_3063099 | 0.07 |
ENST00000221561.8
|
AES
|
amino-terminal enhancer of split |
| chr12_-_54071181 | 0.07 |
ENST00000338662.5
|
ATP5G2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr3_+_145782358 | 0.07 |
ENST00000422482.1
|
AC107021.1
|
HCG1786590; PRO2533; Uncharacterized protein |
| chrX_+_70503037 | 0.07 |
ENST00000535149.1
|
NONO
|
non-POU domain containing, octamer-binding |
| chr17_-_33760164 | 0.07 |
ENST00000445092.1
ENST00000394562.1 ENST00000447040.2 |
SLFN12
|
schlafen family member 12 |
| chr15_+_84904525 | 0.07 |
ENST00000510439.2
|
GOLGA6L4
|
golgin A6 family-like 4 |
| chr7_+_94536898 | 0.07 |
ENST00000433360.1
ENST00000340694.4 ENST00000424654.1 |
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr17_-_34257771 | 0.07 |
ENST00000394529.3
ENST00000293273.6 |
RDM1
|
RAD52 motif 1 |
| chr1_-_153940097 | 0.07 |
ENST00000413622.1
ENST00000310483.6 |
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr7_+_154795154 | 0.07 |
ENST00000608317.1
|
PAXIP1-AS1
|
PAXIP1 antisense RNA 1 (head to head) |
| chr2_+_10861775 | 0.07 |
ENST00000272238.4
ENST00000381661.3 |
ATP6V1C2
|
ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C2 |
| chr13_+_58206655 | 0.07 |
ENST00000377918.3
|
PCDH17
|
protocadherin 17 |
| chr12_-_71148413 | 0.06 |
ENST00000440835.2
ENST00000549308.1 ENST00000550661.1 |
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr7_+_12727250 | 0.06 |
ENST00000404894.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr19_-_42759300 | 0.06 |
ENST00000222329.4
|
ERF
|
Ets2 repressor factor |
| chr2_+_223289208 | 0.06 |
ENST00000321276.7
|
SGPP2
|
sphingosine-1-phosphate phosphatase 2 |
| chr1_-_104239076 | 0.06 |
ENST00000370080.3
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr5_+_138089100 | 0.06 |
ENST00000520339.1
ENST00000355078.5 ENST00000302763.7 ENST00000518910.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr5_+_140430979 | 0.06 |
ENST00000306549.3
|
PCDHB1
|
protocadherin beta 1 |
| chr12_+_53342625 | 0.06 |
ENST00000388837.2
ENST00000550600.1 ENST00000388835.3 |
KRT18
|
keratin 18 |
| chr10_+_26505662 | 0.06 |
ENST00000428517.2
|
GAD2
|
glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) |
| chr2_+_24397930 | 0.06 |
ENST00000295150.3
|
FAM228A
|
family with sequence similarity 228, member A |
| chr17_-_2415169 | 0.06 |
ENST00000263092.6
ENST00000538844.1 ENST00000576976.1 |
METTL16
|
methyltransferase like 16 |
| chr6_+_28092338 | 0.06 |
ENST00000340487.4
|
ZSCAN16
|
zinc finger and SCAN domain containing 16 |
| chr1_-_95392635 | 0.06 |
ENST00000538964.1
ENST00000394202.4 ENST00000370206.4 |
CNN3
|
calponin 3, acidic |
| chr17_+_77704681 | 0.06 |
ENST00000328313.5
|
ENPP7
|
ectonucleotide pyrophosphatase/phosphodiesterase 7 |
| chr12_-_71148357 | 0.06 |
ENST00000378778.1
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr16_+_1031762 | 0.06 |
ENST00000293894.3
|
SOX8
|
SRY (sex determining region Y)-box 8 |
| chr17_-_33760269 | 0.05 |
ENST00000452764.3
|
SLFN12
|
schlafen family member 12 |
| chr12_-_13153139 | 0.05 |
ENST00000536942.1
ENST00000014930.4 |
HEBP1
|
heme binding protein 1 |
| chr1_+_150522222 | 0.05 |
ENST00000369039.5
|
ADAMTSL4
|
ADAMTS-like 4 |
| chr17_-_27405875 | 0.05 |
ENST00000359450.6
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr8_+_110374683 | 0.05 |
ENST00000378402.5
|
PKHD1L1
|
polycystic kidney and hepatic disease 1 (autosomal recessive)-like 1 |
| chr1_-_104238912 | 0.05 |
ENST00000330330.5
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr2_-_175870085 | 0.05 |
ENST00000409156.3
|
CHN1
|
chimerin 1 |
| chr5_+_32710736 | 0.05 |
ENST00000415685.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr1_-_225615599 | 0.05 |
ENST00000421383.1
ENST00000272163.4 |
LBR
|
lamin B receptor |
| chr3_-_143567262 | 0.05 |
ENST00000474151.1
ENST00000316549.6 |
SLC9A9
|
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr5_-_114632307 | 0.05 |
ENST00000506442.1
ENST00000379611.5 |
CCDC112
|
coiled-coil domain containing 112 |
| chr14_+_68086515 | 0.04 |
ENST00000261783.3
|
ARG2
|
arginase 2 |
| chr5_-_127873496 | 0.04 |
ENST00000508989.1
|
FBN2
|
fibrillin 2 |
| chr4_-_89619386 | 0.04 |
ENST00000323061.5
|
NAP1L5
|
nucleosome assembly protein 1-like 5 |
| chr17_+_67410832 | 0.04 |
ENST00000590474.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr1_+_104198377 | 0.04 |
ENST00000370083.4
|
AMY1A
|
amylase, alpha 1A (salivary) |
| chr3_-_114343768 | 0.04 |
ENST00000393785.2
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr1_-_247335269 | 0.04 |
ENST00000543802.2
ENST00000491356.1 ENST00000472531.1 ENST00000340684.6 |
ZNF124
|
zinc finger protein 124 |
| chr1_-_201391149 | 0.04 |
ENST00000555948.1
ENST00000556362.1 |
TNNI1
|
troponin I type 1 (skeletal, slow) |
| chr2_-_24583314 | 0.04 |
ENST00000443927.1
ENST00000406921.3 ENST00000412011.1 |
ITSN2
|
intersectin 2 |
| chr1_+_220701456 | 0.04 |
ENST00000366918.4
ENST00000402574.1 |
MARK1
|
MAP/microtubule affinity-regulating kinase 1 |
| chr3_+_26664291 | 0.04 |
ENST00000396641.2
|
LRRC3B
|
leucine rich repeat containing 3B |
Network of associatons between targets according to the STRING database.
First level regulatory network of SCRT1_SCRT2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:1903285 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.2 | 0.8 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.2 | 0.7 | GO:0042247 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.1 | 0.7 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.1 | 0.9 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.3 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.1 | 0.4 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.1 | 0.2 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.1 | 0.6 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 0.3 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.1 | 0.3 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 0.2 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.3 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.1 | 0.2 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.4 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.2 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 1.0 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.7 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.3 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.3 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.2 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.4 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 1.6 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.1 | GO:0050894 | determination of affect(GO:0050894) |
| 0.0 | 0.1 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.2 | GO:1903401 | lysine transport(GO:0015819) L-lysine transport(GO:1902022) L-lysine transmembrane transport(GO:1903401) |
| 0.0 | 1.3 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.4 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.0 | 0.1 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.0 | 0.3 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.1 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 0.0 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 1.8 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.5 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 0.5 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.5 | GO:0036109 | alpha-linolenic acid metabolic process(GO:0036109) linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.1 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.0 | 0.5 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.0 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.7 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.0 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0043000 | Golgi to plasma membrane CFTR protein transport(GO:0043000) |
| 0.0 | 1.7 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.4 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.9 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.2 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.4 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.4 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.8 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.2 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.2 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.1 | 0.4 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 0.7 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.9 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.4 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 1.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 1.0 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 1.0 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 1.8 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.5 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 0.3 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.1 | 0.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.2 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.9 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 0.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.9 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.0 | 0.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 1.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.2 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.5 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.0 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.4 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.4 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.0 | GO:0016160 | amylase activity(GO:0016160) |
| 0.0 | 0.1 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |