Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
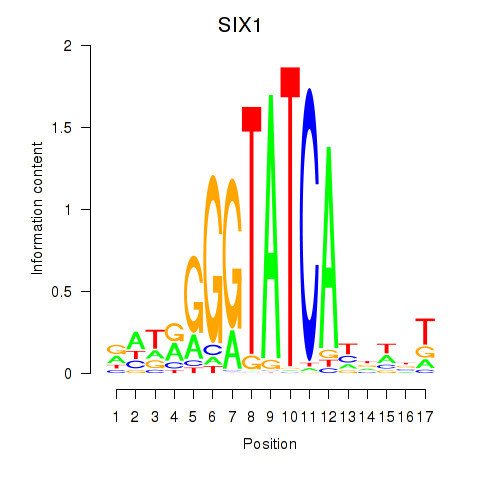
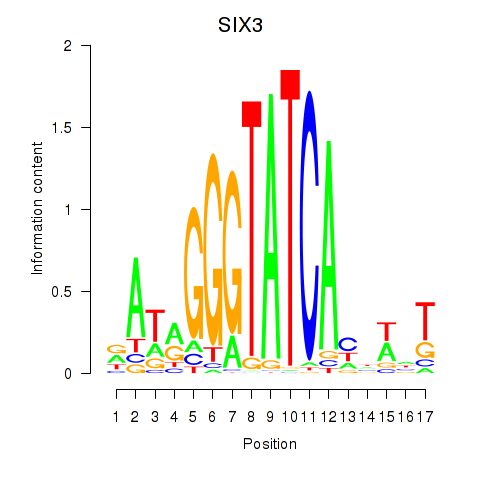
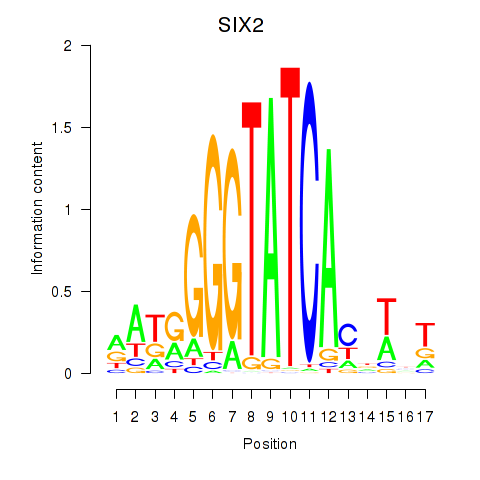
Results for SIX1_SIX3_SIX2
Z-value: 0.72
Transcription factors associated with SIX1_SIX3_SIX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SIX1
|
ENSG00000126778.7 | SIX homeobox 1 |
|
SIX3
|
ENSG00000138083.3 | SIX homeobox 3 |
|
SIX2
|
ENSG00000170577.7 | SIX homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SIX2 | hg19_v2_chr2_-_45236540_45236577 | 0.21 | 3.2e-01 | Click! |
| SIX1 | hg19_v2_chr14_-_61124977_61125037 | 0.20 | 3.4e-01 | Click! |
| SIX3 | hg19_v2_chr2_+_45168875_45168916 | 0.03 | 9.0e-01 | Click! |
Activity profile of SIX1_SIX3_SIX2 motif
Sorted Z-values of SIX1_SIX3_SIX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_160166218 | 2.24 |
ENST00000537657.1
|
SOD2
|
superoxide dismutase 2, mitochondrial |
| chr5_+_125758813 | 2.13 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr5_+_125758865 | 2.09 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr21_+_27011899 | 1.77 |
ENST00000425221.2
|
JAM2
|
junctional adhesion molecule 2 |
| chr6_-_154751629 | 1.71 |
ENST00000424998.1
|
CNKSR3
|
CNKSR family member 3 |
| chr10_-_49860525 | 1.27 |
ENST00000435790.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr21_+_27011584 | 1.24 |
ENST00000400532.1
ENST00000480456.1 ENST00000312957.5 |
JAM2
|
junctional adhesion molecule 2 |
| chr1_-_8000872 | 1.06 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr6_-_136847099 | 0.92 |
ENST00000438100.2
|
MAP7
|
microtubule-associated protein 7 |
| chr6_+_31554779 | 0.89 |
ENST00000376090.2
|
LST1
|
leukocyte specific transcript 1 |
| chr19_-_56249740 | 0.89 |
ENST00000590200.1
ENST00000332836.2 |
NLRP9
|
NLR family, pyrin domain containing 9 |
| chr3_-_11685345 | 0.86 |
ENST00000430365.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr1_+_26737253 | 0.82 |
ENST00000326279.6
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr15_-_55562582 | 0.76 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr1_-_240906911 | 0.74 |
ENST00000431139.2
|
RP11-80B9.4
|
RP11-80B9.4 |
| chr6_-_136847610 | 0.73 |
ENST00000454590.1
ENST00000432797.2 |
MAP7
|
microtubule-associated protein 7 |
| chr6_+_31554962 | 0.72 |
ENST00000376092.3
ENST00000376086.3 ENST00000303757.8 ENST00000376093.2 ENST00000376102.3 |
LST1
|
leukocyte specific transcript 1 |
| chr1_+_12916941 | 0.69 |
ENST00000240189.2
|
PRAMEF2
|
PRAME family member 2 |
| chr16_+_56642041 | 0.64 |
ENST00000245185.5
|
MT2A
|
metallothionein 2A |
| chr1_+_26737292 | 0.61 |
ENST00000254231.4
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr13_-_46679185 | 0.59 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr11_+_34654011 | 0.58 |
ENST00000531794.1
|
EHF
|
ets homologous factor |
| chr13_-_46679144 | 0.58 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr18_+_55816546 | 0.57 |
ENST00000435432.2
ENST00000357895.5 ENST00000586263.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr1_-_151778630 | 0.57 |
ENST00000368820.3
|
LINGO4
|
leucine rich repeat and Ig domain containing 4 |
| chr12_+_49121213 | 0.56 |
ENST00000548380.1
|
LINC00935
|
long intergenic non-protein coding RNA 935 |
| chr6_+_31554456 | 0.56 |
ENST00000339530.4
|
LST1
|
leukocyte specific transcript 1 |
| chr11_-_34533257 | 0.55 |
ENST00000312319.2
|
ELF5
|
E74-like factor 5 (ets domain transcription factor) |
| chrX_+_144908928 | 0.54 |
ENST00000408967.2
|
TMEM257
|
transmembrane protein 257 |
| chrX_+_56259316 | 0.53 |
ENST00000468660.1
|
KLF8
|
Kruppel-like factor 8 |
| chr16_+_56666563 | 0.52 |
ENST00000570233.1
|
MT1M
|
metallothionein 1M |
| chr1_+_145293371 | 0.51 |
ENST00000342960.5
|
NBPF10
|
neuroblastoma breakpoint family, member 10 |
| chr2_+_161993465 | 0.50 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr11_+_64323098 | 0.48 |
ENST00000301891.4
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr4_+_89299994 | 0.48 |
ENST00000264346.7
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr1_-_47082495 | 0.48 |
ENST00000545730.1
ENST00000531769.1 ENST00000319928.3 |
MKNK1
MOB3C
|
MAP kinase interacting serine/threonine kinase 1 MOB kinase activator 3C |
| chr16_+_57673207 | 0.47 |
ENST00000564783.1
ENST00000564729.1 ENST00000565976.1 ENST00000566508.1 ENST00000544297.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr17_+_6659153 | 0.47 |
ENST00000441631.1
ENST00000438512.1 ENST00000346752.4 ENST00000361842.3 |
XAF1
|
XIAP associated factor 1 |
| chr6_-_138428613 | 0.46 |
ENST00000421351.3
|
PERP
|
PERP, TP53 apoptosis effector |
| chr11_+_64323428 | 0.46 |
ENST00000377581.3
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr11_+_64323156 | 0.46 |
ENST00000377585.3
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr1_-_13115578 | 0.46 |
ENST00000414205.2
|
PRAMEF6
|
PRAME family member 6 |
| chrX_+_101470280 | 0.44 |
ENST00000395088.2
ENST00000330252.5 ENST00000333110.5 |
NXF2
TCP11X1
|
nuclear RNA export factor 2 t-complex 11 family, X-linked 1 |
| chr1_+_12851545 | 0.44 |
ENST00000332296.7
|
PRAMEF1
|
PRAME family member 1 |
| chr1_-_13390765 | 0.43 |
ENST00000357367.2
|
PRAMEF8
|
PRAME family member 8 |
| chr14_-_24615805 | 0.43 |
ENST00000560410.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr19_+_50270219 | 0.42 |
ENST00000354293.5
ENST00000359032.5 |
AP2A1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr9_-_117568365 | 0.42 |
ENST00000374045.4
|
TNFSF15
|
tumor necrosis factor (ligand) superfamily, member 15 |
| chr19_+_11457232 | 0.41 |
ENST00000587531.1
|
CCDC159
|
coiled-coil domain containing 159 |
| chr8_-_134072593 | 0.41 |
ENST00000427060.2
|
SLA
|
Src-like-adaptor |
| chr20_+_34824355 | 0.40 |
ENST00000397286.3
ENST00000320849.4 ENST00000373932.3 |
AAR2
|
AAR2 splicing factor homolog (S. cerevisiae) |
| chr11_-_32452357 | 0.40 |
ENST00000379079.2
ENST00000530998.1 |
WT1
|
Wilms tumor 1 |
| chr12_-_89920030 | 0.39 |
ENST00000413530.1
ENST00000547474.1 |
GALNT4
POC1B-GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) POC1B-GALNT4 readthrough |
| chr3_+_111718036 | 0.39 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr1_-_13117736 | 0.38 |
ENST00000376192.5
ENST00000376182.1 |
PRAMEF6
|
PRAME family member 6 |
| chr3_+_111717600 | 0.37 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr14_+_58711539 | 0.37 |
ENST00000216455.4
ENST00000412908.2 ENST00000557508.1 |
PSMA3
|
proteasome (prosome, macropain) subunit, alpha type, 3 |
| chr17_+_7465216 | 0.37 |
ENST00000321337.7
|
SENP3
|
SUMO1/sentrin/SMT3 specific peptidase 3 |
| chr5_-_133304473 | 0.36 |
ENST00000231512.3
|
C5orf15
|
chromosome 5 open reading frame 15 |
| chr8_+_18248755 | 0.35 |
ENST00000286479.3
|
NAT2
|
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
| chr8_+_18248786 | 0.35 |
ENST00000520116.1
|
NAT2
|
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
| chr19_-_58446721 | 0.35 |
ENST00000396147.1
ENST00000595569.1 ENST00000599852.1 ENST00000425570.3 ENST00000601593.1 |
ZNF418
|
zinc finger protein 418 |
| chr12_-_89919965 | 0.35 |
ENST00000548729.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr8_-_48651648 | 0.35 |
ENST00000408965.3
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr2_+_161993412 | 0.33 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr7_-_75452673 | 0.33 |
ENST00000416943.1
|
CCL24
|
chemokine (C-C motif) ligand 24 |
| chr11_-_58499434 | 0.32 |
ENST00000344743.3
ENST00000278400.3 |
GLYAT
|
glycine-N-acyltransferase |
| chr9_+_12693336 | 0.32 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr8_-_134115118 | 0.31 |
ENST00000395352.3
ENST00000338087.5 |
SLA
|
Src-like-adaptor |
| chr1_+_35225339 | 0.31 |
ENST00000339480.1
|
GJB4
|
gap junction protein, beta 4, 30.3kDa |
| chr8_+_99129513 | 0.31 |
ENST00000522319.1
ENST00000401707.2 |
POP1
|
processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr1_+_13359819 | 0.31 |
ENST00000376168.1
|
PRAMEF5
|
PRAME family member 5 |
| chr6_+_31555045 | 0.31 |
ENST00000396101.3
ENST00000490742.1 |
LST1
|
leukocyte specific transcript 1 |
| chr11_-_5364809 | 0.30 |
ENST00000300773.2
|
OR51B5
|
olfactory receptor, family 51, subfamily B, member 5 |
| chr12_-_323689 | 0.30 |
ENST00000428720.1
|
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr14_+_51026743 | 0.30 |
ENST00000358385.6
ENST00000357032.3 ENST00000354525.4 |
ATL1
|
atlastin GTPase 1 |
| chr22_-_43091639 | 0.29 |
ENST00000249005.2
ENST00000381278.3 |
A4GALT
|
alpha 1,4-galactosyltransferase |
| chr2_-_219134822 | 0.29 |
ENST00000444053.1
ENST00000248450.4 |
AAMP
|
angio-associated, migratory cell protein |
| chr3_-_157251383 | 0.28 |
ENST00000487753.1
ENST00000489602.1 ENST00000461299.1 ENST00000479987.1 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr4_+_70894130 | 0.27 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr11_+_70244510 | 0.27 |
ENST00000346329.3
ENST00000301843.8 ENST00000376561.3 |
CTTN
|
cortactin |
| chr12_+_20848377 | 0.27 |
ENST00000540354.1
ENST00000266509.2 ENST00000381552.1 |
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr21_+_35736302 | 0.26 |
ENST00000290310.3
|
KCNE2
|
potassium voltage-gated channel, Isk-related family, member 2 |
| chr7_-_44198874 | 0.26 |
ENST00000395796.3
ENST00000345378.2 |
GCK
|
glucokinase (hexokinase 4) |
| chr1_+_87595542 | 0.26 |
ENST00000461990.1
|
RP5-1052I5.1
|
long intergenic non-protein coding RNA 1140 |
| chr19_-_11039261 | 0.26 |
ENST00000590329.1
ENST00000587943.1 ENST00000585858.1 ENST00000586748.1 ENST00000586575.1 ENST00000253031.2 |
YIPF2
|
Yip1 domain family, member 2 |
| chr2_-_89266286 | 0.26 |
ENST00000464162.1
|
IGKV1-6
|
immunoglobulin kappa variable 1-6 |
| chr12_-_9913489 | 0.26 |
ENST00000228434.3
ENST00000536709.1 |
CD69
|
CD69 molecule |
| chrX_+_13587712 | 0.25 |
ENST00000361306.1
ENST00000380602.3 |
EGFL6
|
EGF-like-domain, multiple 6 |
| chr15_+_75491213 | 0.25 |
ENST00000360639.2
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr17_+_46184911 | 0.25 |
ENST00000580219.1
ENST00000452859.2 ENST00000393405.2 ENST00000439357.2 ENST00000359238.2 |
SNX11
|
sorting nexin 11 |
| chr6_+_88757507 | 0.25 |
ENST00000237201.1
|
SPACA1
|
sperm acrosome associated 1 |
| chr9_-_2844058 | 0.24 |
ENST00000397885.2
|
KIAA0020
|
KIAA0020 |
| chr3_-_10452359 | 0.23 |
ENST00000452124.1
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr19_-_55549624 | 0.23 |
ENST00000417454.1
ENST00000310373.3 ENST00000333884.2 |
GP6
|
glycoprotein VI (platelet) |
| chr3_+_70048881 | 0.23 |
ENST00000483525.1
|
RP11-460N16.1
|
RP11-460N16.1 |
| chr7_+_117251671 | 0.23 |
ENST00000468795.1
|
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr6_-_167797887 | 0.23 |
ENST00000476779.2
ENST00000460930.2 ENST00000397829.4 ENST00000366827.2 |
TCP10
|
t-complex 10 |
| chr1_+_169337172 | 0.23 |
ENST00000367807.3
ENST00000367808.3 ENST00000329281.2 ENST00000420531.1 |
BLZF1
|
basic leucine zipper nuclear factor 1 |
| chr3_+_42850959 | 0.23 |
ENST00000442925.1
ENST00000422265.1 ENST00000497921.1 ENST00000426937.1 |
ACKR2
KRBOX1
|
atypical chemokine receptor 2 KRAB box domain containing 1 |
| chr16_-_67867749 | 0.22 |
ENST00000566758.1
ENST00000445712.2 ENST00000219172.3 ENST00000564817.1 |
CENPT
|
centromere protein T |
| chr1_+_145293114 | 0.22 |
ENST00000369338.1
|
NBPF10
|
neuroblastoma breakpoint family, member 10 |
| chr2_-_152146385 | 0.22 |
ENST00000414946.1
ENST00000243346.5 |
NMI
|
N-myc (and STAT) interactor |
| chr11_-_5173599 | 0.22 |
ENST00000328942.1
|
OR52A1
|
olfactory receptor, family 52, subfamily A, member 1 |
| chr6_-_11779174 | 0.22 |
ENST00000379413.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr9_+_132427883 | 0.22 |
ENST00000372469.4
|
PRRX2
|
paired related homeobox 2 |
| chr17_+_32646055 | 0.21 |
ENST00000394620.1
|
CCL8
|
chemokine (C-C motif) ligand 8 |
| chr2_-_89597542 | 0.21 |
ENST00000465170.1
|
IGKV1-37
|
immunoglobulin kappa variable 1-37 (non-functional) |
| chr16_+_58010339 | 0.21 |
ENST00000290871.5
ENST00000441824.2 |
TEPP
|
testis, prostate and placenta expressed |
| chr5_-_59481406 | 0.21 |
ENST00000546160.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr5_-_132073210 | 0.21 |
ENST00000378735.1
ENST00000378746.4 |
KIF3A
|
kinesin family member 3A |
| chr2_-_89513402 | 0.21 |
ENST00000498435.1
|
IGKV1-27
|
immunoglobulin kappa variable 1-27 |
| chr15_-_72523454 | 0.20 |
ENST00000565154.1
ENST00000565184.1 ENST00000389093.3 ENST00000449901.2 ENST00000335181.5 ENST00000319622.6 |
PKM
|
pyruvate kinase, muscle |
| chr19_+_41594377 | 0.20 |
ENST00000330436.3
|
CYP2A13
|
cytochrome P450, family 2, subfamily A, polypeptide 13 |
| chr6_-_11779014 | 0.20 |
ENST00000229583.5
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr1_-_12946025 | 0.20 |
ENST00000235349.5
|
PRAMEF4
|
PRAME family member 4 |
| chr7_-_14880892 | 0.20 |
ENST00000406247.3
ENST00000399322.3 ENST00000258767.5 |
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr3_+_149191723 | 0.20 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr1_-_220220000 | 0.20 |
ENST00000366923.3
|
EPRS
|
glutamyl-prolyl-tRNA synthetase |
| chr15_+_89631647 | 0.20 |
ENST00000569550.1
ENST00000565066.1 ENST00000565973.1 |
ABHD2
|
abhydrolase domain containing 2 |
| chr1_-_13007420 | 0.20 |
ENST00000376189.1
|
PRAMEF6
|
PRAME family member 6 |
| chr3_+_178253993 | 0.20 |
ENST00000420517.2
ENST00000452583.1 |
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr12_+_57624119 | 0.19 |
ENST00000555773.1
ENST00000554975.1 ENST00000449049.3 ENST00000393827.4 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr16_+_31119615 | 0.19 |
ENST00000394950.3
ENST00000287507.3 ENST00000219794.6 ENST00000561755.1 |
BCKDK
|
branched chain ketoacid dehydrogenase kinase |
| chr19_+_49109990 | 0.19 |
ENST00000321762.1
|
SPACA4
|
sperm acrosome associated 4 |
| chr21_-_34185944 | 0.19 |
ENST00000479548.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr6_+_31554612 | 0.19 |
ENST00000211921.7
|
LST1
|
leukocyte specific transcript 1 |
| chr17_+_6915798 | 0.19 |
ENST00000402093.1
|
RNASEK
|
ribonuclease, RNase K |
| chr6_+_131894284 | 0.19 |
ENST00000368087.3
ENST00000356962.2 |
ARG1
|
arginase 1 |
| chr16_+_21695750 | 0.18 |
ENST00000388956.4
|
OTOA
|
otoancorin |
| chr20_-_55100981 | 0.18 |
ENST00000243913.4
|
GCNT7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr11_+_55594695 | 0.18 |
ENST00000378397.1
|
OR5L2
|
olfactory receptor, family 5, subfamily L, member 2 |
| chr1_+_12976450 | 0.18 |
ENST00000361079.2
|
PRAMEF7
|
PRAME family member 7 |
| chr6_-_31509714 | 0.18 |
ENST00000456662.1
ENST00000431908.1 ENST00000456976.1 ENST00000428450.1 ENST00000453105.2 ENST00000418897.1 ENST00000415382.2 ENST00000449074.2 ENST00000419020.1 ENST00000428098.1 |
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr6_+_31554636 | 0.18 |
ENST00000433492.1
|
LST1
|
leukocyte specific transcript 1 |
| chr3_+_127771212 | 0.18 |
ENST00000243253.3
ENST00000481210.1 |
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr7_+_100551239 | 0.18 |
ENST00000319509.7
|
MUC3A
|
mucin 3A, cell surface associated |
| chr7_+_75511362 | 0.17 |
ENST00000428119.1
|
RHBDD2
|
rhomboid domain containing 2 |
| chr4_-_156297949 | 0.17 |
ENST00000515654.1
|
MAP9
|
microtubule-associated protein 9 |
| chr4_-_153332886 | 0.17 |
ENST00000603841.1
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr1_-_109618566 | 0.17 |
ENST00000338366.5
|
TAF13
|
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa |
| chr10_-_96122682 | 0.17 |
ENST00000371361.3
|
NOC3L
|
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr14_-_24911448 | 0.17 |
ENST00000555355.1
ENST00000553343.1 ENST00000556523.1 ENST00000556249.1 ENST00000538105.2 ENST00000555225.1 |
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr12_+_110011571 | 0.17 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr15_+_81426588 | 0.17 |
ENST00000286732.4
|
C15orf26
|
chromosome 15 open reading frame 26 |
| chrX_-_65259914 | 0.16 |
ENST00000374737.4
ENST00000455586.2 |
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr3_+_38323785 | 0.16 |
ENST00000466887.1
ENST00000448498.1 |
SLC22A14
|
solute carrier family 22, member 14 |
| chr1_-_108743471 | 0.16 |
ENST00000569674.1
|
SLC25A24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr19_+_11039391 | 0.16 |
ENST00000270502.6
|
C19orf52
|
chromosome 19 open reading frame 52 |
| chr19_+_2867325 | 0.16 |
ENST00000307635.2
ENST00000586426.1 |
ZNF556
|
zinc finger protein 556 |
| chr18_+_616672 | 0.15 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chrX_-_65259900 | 0.15 |
ENST00000412866.2
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr10_-_103603677 | 0.15 |
ENST00000358038.3
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr6_-_43655511 | 0.15 |
ENST00000372133.3
ENST00000372116.1 ENST00000427312.1 |
MRPS18A
|
mitochondrial ribosomal protein S18A |
| chr15_+_62853562 | 0.15 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr16_+_50059125 | 0.15 |
ENST00000427478.2
|
CNEP1R1
|
CTD nuclear envelope phosphatase 1 regulatory subunit 1 |
| chr1_-_11115877 | 0.15 |
ENST00000490101.1
|
SRM
|
spermidine synthase |
| chr2_-_207630033 | 0.15 |
ENST00000449792.1
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
| chr17_+_6915902 | 0.14 |
ENST00000570898.1
ENST00000552842.1 |
RNASEK
|
ribonuclease, RNase K |
| chr13_+_28712614 | 0.14 |
ENST00000380958.3
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr6_+_30035307 | 0.14 |
ENST00000376765.2
ENST00000376763.1 |
PPP1R11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chr19_+_16222439 | 0.14 |
ENST00000300935.3
|
RAB8A
|
RAB8A, member RAS oncogene family |
| chr10_+_7745303 | 0.14 |
ENST00000429820.1
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr17_+_6915730 | 0.14 |
ENST00000548577.1
|
RNASEK
|
ribonuclease, RNase K |
| chr18_+_19192228 | 0.14 |
ENST00000300413.5
ENST00000579618.1 ENST00000582475.1 |
SNRPD1
|
small nuclear ribonucleoprotein D1 polypeptide 16kDa |
| chr4_+_155484155 | 0.14 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr2_-_114514181 | 0.14 |
ENST00000409342.1
|
SLC35F5
|
solute carrier family 35, member F5 |
| chr12_-_58135903 | 0.14 |
ENST00000257897.3
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr10_+_135207623 | 0.14 |
ENST00000317502.6
ENST00000432508.3 |
MTG1
|
mitochondrial ribosome-associated GTPase 1 |
| chr19_+_17420340 | 0.14 |
ENST00000359866.4
|
DDA1
|
DET1 and DDB1 associated 1 |
| chr13_-_60737898 | 0.13 |
ENST00000377908.2
ENST00000400319.1 ENST00000400320.1 ENST00000267215.4 |
DIAPH3
|
diaphanous-related formin 3 |
| chr17_-_3030875 | 0.13 |
ENST00000328890.2
|
OR1G1
|
olfactory receptor, family 1, subfamily G, member 1 |
| chr11_-_104840093 | 0.13 |
ENST00000417440.2
ENST00000444739.2 |
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr2_+_89923550 | 0.13 |
ENST00000509129.1
|
IGKV1D-37
|
immunoglobulin kappa variable 1D-37 (non-functional) |
| chrX_-_72095808 | 0.13 |
ENST00000373529.5
|
DMRTC1
|
DMRT-like family C1 |
| chr10_+_135207598 | 0.13 |
ENST00000477902.2
|
MTG1
|
mitochondrial ribosome-associated GTPase 1 |
| chr1_-_166845515 | 0.13 |
ENST00000367874.4
|
TADA1
|
transcriptional adaptor 1 |
| chr1_+_196946664 | 0.13 |
ENST00000367414.5
|
CFHR5
|
complement factor H-related 5 |
| chr20_+_18488137 | 0.13 |
ENST00000450074.1
ENST00000262544.2 ENST00000336714.3 ENST00000377475.3 |
SEC23B
|
Sec23 homolog B (S. cerevisiae) |
| chr5_-_145562147 | 0.13 |
ENST00000545646.1
ENST00000274562.9 ENST00000510191.1 ENST00000394434.2 |
LARS
|
leucyl-tRNA synthetase |
| chr17_-_8151353 | 0.13 |
ENST00000315684.8
|
CTC1
|
CTS telomere maintenance complex component 1 |
| chr10_-_74856608 | 0.13 |
ENST00000307116.2
ENST00000373008.2 ENST00000412021.2 ENST00000394890.2 ENST00000263556.3 ENST00000440381.1 |
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I |
| chr11_+_71791359 | 0.13 |
ENST00000419228.1
ENST00000435085.1 ENST00000307198.7 ENST00000538413.1 |
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr7_-_48068699 | 0.13 |
ENST00000412142.1
ENST00000395572.2 |
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr8_+_118147498 | 0.12 |
ENST00000519688.1
ENST00000456015.2 |
SLC30A8
|
solute carrier family 30 (zinc transporter), member 8 |
| chr2_+_183989157 | 0.12 |
ENST00000541912.1
|
NUP35
|
nucleoporin 35kDa |
| chr17_+_39240459 | 0.12 |
ENST00000391417.4
|
KRTAP4-7
|
keratin associated protein 4-7 |
| chr17_+_7210294 | 0.12 |
ENST00000336452.7
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr12_-_54758251 | 0.12 |
ENST00000267015.3
ENST00000551809.1 |
GPR84
|
G protein-coupled receptor 84 |
| chr12_+_26205496 | 0.12 |
ENST00000537946.1
ENST00000541218.1 ENST00000282884.9 ENST00000545413.1 |
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr10_-_103603568 | 0.12 |
ENST00000356640.2
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr12_+_81471816 | 0.12 |
ENST00000261206.3
|
ACSS3
|
acyl-CoA synthetase short-chain family member 3 |
| chr6_+_31583761 | 0.12 |
ENST00000376049.4
|
AIF1
|
allograft inflammatory factor 1 |
| chr14_-_24740709 | 0.12 |
ENST00000399409.3
ENST00000216840.6 |
RABGGTA
|
Rab geranylgeranyltransferase, alpha subunit |
| chr11_+_114168773 | 0.12 |
ENST00000542647.1
ENST00000545255.1 |
NNMT
|
nicotinamide N-methyltransferase |
| chr7_+_123241908 | 0.12 |
ENST00000434204.1
ENST00000437535.1 ENST00000451215.1 |
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr8_-_25902876 | 0.11 |
ENST00000520164.1
|
EBF2
|
early B-cell factor 2 |
| chr2_-_105953912 | 0.11 |
ENST00000610036.1
|
RP11-332H14.2
|
RP11-332H14.2 |
| chr12_+_20848486 | 0.11 |
ENST00000545102.1
|
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr2_-_207629997 | 0.11 |
ENST00000454776.2
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
Network of associatons between targets according to the STRING database.
First level regulatory network of SIX1_SIX3_SIX2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0001315 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.3 | 1.2 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.2 | 1.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.2 | 1.1 | GO:2001183 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.1 | 0.4 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 | 0.8 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.6 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.1 | 0.8 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.5 | GO:1902159 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.1 | 0.6 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 1.4 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.4 | GO:0072299 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.1 | 0.5 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 0.2 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.1 | 0.2 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.3 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.1 | 0.2 | GO:0046833 | snRNA export from nucleus(GO:0006408) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 1.7 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.3 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.0 | 0.3 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.0 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.3 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.3 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.0 | 0.2 | GO:0045354 | negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.0 | 0.2 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.0 | 0.3 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.5 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.2 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.0 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.4 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.2 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.1 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.0 | 0.3 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 3.1 | GO:0050672 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) |
| 0.0 | 0.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.0 | 0.2 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.4 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0090299 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.0 | 0.2 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.1 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.3 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.3 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.3 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.4 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.2 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.2 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.5 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.1 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.0 | 0.0 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.0 | 0.1 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.3 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.3 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.7 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.0 | 0.3 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.0 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.2 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.5 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.0 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.4 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 0.2 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.1 | 0.8 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.2 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.2 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.3 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.3 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.4 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.3 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.1 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.0 | 1.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 2.9 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 0.7 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 0.8 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 1.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 1.4 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.4 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 0.2 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 0.2 | GO:0004818 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.1 | 0.3 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 0.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.2 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.6 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.3 | GO:0004793 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.6 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.0 | 0.2 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.1 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.3 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 1.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.4 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 1.1 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.2 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.3 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.0 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.0 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.9 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 2.2 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.1 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.5 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 3.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.2 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.6 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.8 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |