Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
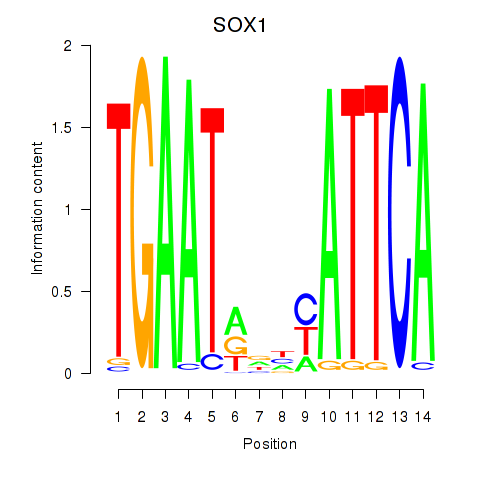
Results for SOX1
Z-value: 0.83
Transcription factors associated with SOX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX1
|
ENSG00000182968.3 | SRY-box transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX1 | hg19_v2_chr13_+_112721913_112721913 | -0.02 | 9.2e-01 | Click! |
Activity profile of SOX1 motif
Sorted Z-values of SOX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_154622652 | 3.53 |
ENST00000260010.6
|
TLR2
|
toll-like receptor 2 |
| chr5_-_150460914 | 2.73 |
ENST00000389378.2
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr19_-_10491234 | 2.52 |
ENST00000524462.1
ENST00000531836.1 ENST00000525621.1 |
TYK2
|
tyrosine kinase 2 |
| chr1_+_65613340 | 1.82 |
ENST00000546702.1
|
AK4
|
adenylate kinase 4 |
| chr3_-_189840223 | 1.68 |
ENST00000427335.2
|
LEPREL1
|
leprecan-like 1 |
| chr11_-_57177586 | 1.59 |
ENST00000529411.1
|
RP11-872D17.8
|
Uncharacterized protein |
| chr5_-_16916624 | 1.35 |
ENST00000513882.1
|
MYO10
|
myosin X |
| chr19_+_3762645 | 1.20 |
ENST00000330133.4
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr18_+_21719018 | 1.19 |
ENST00000585037.1
ENST00000415309.2 ENST00000399481.2 ENST00000577705.1 ENST00000327201.6 |
CABYR
|
calcium binding tyrosine-(Y)-phosphorylation regulated |
| chr1_+_65613217 | 1.14 |
ENST00000545314.1
|
AK4
|
adenylate kinase 4 |
| chr2_-_163175133 | 1.11 |
ENST00000421365.2
ENST00000263642.2 |
IFIH1
|
interferon induced with helicase C domain 1 |
| chrX_+_11129388 | 1.06 |
ENST00000321143.4
ENST00000380763.3 ENST00000380762.4 |
HCCS
|
holocytochrome c synthase |
| chr6_+_149539767 | 1.06 |
ENST00000606202.1
ENST00000536230.1 ENST00000445901.1 |
TAB2
RP1-111D6.3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 RP1-111D6.3 |
| chr20_+_31805131 | 0.98 |
ENST00000375454.3
ENST00000375452.3 |
BPIFA3
|
BPI fold containing family A, member 3 |
| chr2_+_234296792 | 0.97 |
ENST00000409813.3
|
DGKD
|
diacylglycerol kinase, delta 130kDa |
| chr12_+_75874460 | 0.92 |
ENST00000266659.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr1_-_237167718 | 0.90 |
ENST00000464121.2
|
MT1HL1
|
metallothionein 1H-like 1 |
| chr3_-_16524357 | 0.87 |
ENST00000432519.1
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr9_-_85882145 | 0.86 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr3_-_58613323 | 0.79 |
ENST00000474531.1
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107, member A |
| chr10_-_92681033 | 0.79 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr9_-_2844058 | 0.73 |
ENST00000397885.2
|
KIAA0020
|
KIAA0020 |
| chr19_+_21324827 | 0.71 |
ENST00000600692.1
ENST00000599296.1 ENST00000594425.1 ENST00000311048.7 |
ZNF431
|
zinc finger protein 431 |
| chr1_+_46805832 | 0.70 |
ENST00000474844.1
|
NSUN4
|
NOP2/Sun domain family, member 4 |
| chr14_-_25078864 | 0.69 |
ENST00000216338.4
ENST00000557220.2 ENST00000382548.4 |
GZMH
|
granzyme H (cathepsin G-like 2, protein h-CCPX) |
| chr14_-_74417096 | 0.68 |
ENST00000286544.3
|
FAM161B
|
family with sequence similarity 161, member B |
| chr19_+_3762703 | 0.66 |
ENST00000589174.1
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr12_+_9980069 | 0.65 |
ENST00000354855.3
ENST00000324214.4 ENST00000279544.3 |
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr1_+_19578033 | 0.63 |
ENST00000330263.4
|
MRTO4
|
mRNA turnover 4 homolog (S. cerevisiae) |
| chr11_+_55029628 | 0.61 |
ENST00000417545.2
|
TRIM48
|
tripartite motif containing 48 |
| chr11_-_133715394 | 0.59 |
ENST00000299140.3
ENST00000532889.1 |
SPATA19
|
spermatogenesis associated 19 |
| chr11_-_113577052 | 0.58 |
ENST00000540540.1
ENST00000545579.1 ENST00000538955.1 ENST00000299882.5 |
TMPRSS5
|
transmembrane protease, serine 5 |
| chr1_+_144811943 | 0.57 |
ENST00000281815.8
|
NBPF9
|
neuroblastoma breakpoint family, member 9 |
| chr11_-_104827425 | 0.57 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr2_-_209054709 | 0.56 |
ENST00000449053.1
ENST00000451346.1 ENST00000341287.4 |
C2orf80
|
chromosome 2 open reading frame 80 |
| chr4_-_170924888 | 0.56 |
ENST00000502832.1
ENST00000393704.3 |
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr13_+_109248500 | 0.55 |
ENST00000356711.2
|
MYO16
|
myosin XVI |
| chr19_+_17830051 | 0.54 |
ENST00000594625.1
ENST00000324096.4 ENST00000600186.1 ENST00000597735.1 |
MAP1S
|
microtubule-associated protein 1S |
| chr10_-_73976025 | 0.53 |
ENST00000342444.4
ENST00000533958.1 ENST00000527593.1 ENST00000394915.3 ENST00000530461.1 ENST00000317168.6 ENST00000524829.1 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr4_+_156680153 | 0.52 |
ENST00000502959.1
ENST00000505764.1 ENST00000507146.1 ENST00000264424.8 ENST00000503520.1 |
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr1_+_148560843 | 0.52 |
ENST00000442702.2
ENST00000369187.3 |
NBPF15
|
neuroblastoma breakpoint family, member 15 |
| chr20_-_14318248 | 0.52 |
ENST00000378053.3
ENST00000341420.4 |
FLRT3
|
fibronectin leucine rich transmembrane protein 3 |
| chr11_-_113577014 | 0.52 |
ENST00000544634.1
ENST00000539732.1 ENST00000538770.1 ENST00000536856.1 ENST00000544476.1 |
TMPRSS5
|
transmembrane protease, serine 5 |
| chr13_+_49280951 | 0.51 |
ENST00000282018.3
|
CYSLTR2
|
cysteinyl leukotriene receptor 2 |
| chr11_-_123756334 | 0.50 |
ENST00000528595.1
ENST00000375026.2 |
TMEM225
|
transmembrane protein 225 |
| chr1_-_52499443 | 0.48 |
ENST00000371614.1
|
KTI12
|
KTI12 homolog, chromatin associated (S. cerevisiae) |
| chr12_-_127174845 | 0.48 |
ENST00000540814.1
ENST00000541065.1 ENST00000541769.1 |
RP11-407A16.3
RP11-407A16.4
|
RP11-407A16.3 RP11-407A16.4 |
| chr17_-_29624343 | 0.46 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr17_+_33448593 | 0.46 |
ENST00000158009.5
|
FNDC8
|
fibronectin type III domain containing 8 |
| chr1_-_200379180 | 0.44 |
ENST00000294740.3
|
ZNF281
|
zinc finger protein 281 |
| chr11_-_47400078 | 0.42 |
ENST00000378538.3
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr15_+_59903975 | 0.42 |
ENST00000560585.1
ENST00000396065.1 |
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr2_+_11864458 | 0.42 |
ENST00000396098.1
ENST00000396099.1 ENST00000425416.2 |
LPIN1
|
lipin 1 |
| chr20_+_49575342 | 0.41 |
ENST00000244051.1
|
MOCS3
|
molybdenum cofactor synthesis 3 |
| chr1_+_174769006 | 0.41 |
ENST00000489615.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr10_+_15085895 | 0.41 |
ENST00000378228.3
|
OLAH
|
oleoyl-ACP hydrolase |
| chr12_-_10151773 | 0.40 |
ENST00000298527.6
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1, member B |
| chr7_-_142176790 | 0.40 |
ENST00000390369.2
|
TRBV7-4
|
T cell receptor beta variable 7-4 (gene/pseudogene) |
| chr6_+_123317116 | 0.40 |
ENST00000275162.5
|
CLVS2
|
clavesin 2 |
| chr20_-_23586513 | 0.40 |
ENST00000376971.3
|
CST9
|
cystatin 9 (testatin) |
| chr3_+_138327542 | 0.39 |
ENST00000360570.3
ENST00000393035.2 |
FAIM
|
Fas apoptotic inhibitory molecule |
| chr10_-_43892668 | 0.38 |
ENST00000544000.1
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr3_+_138327417 | 0.38 |
ENST00000338446.4
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr11_-_47399942 | 0.37 |
ENST00000227163.4
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr3_+_127770455 | 0.37 |
ENST00000464451.1
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr10_-_73976884 | 0.35 |
ENST00000317126.4
ENST00000545550.1 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr12_+_115800817 | 0.35 |
ENST00000547948.1
|
RP11-116D17.1
|
HCG2038717; Uncharacterized protein |
| chr11_-_47400062 | 0.34 |
ENST00000533030.1
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr1_-_43638168 | 0.33 |
ENST00000431635.2
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chr6_-_130543958 | 0.32 |
ENST00000437477.2
ENST00000439090.2 |
SAMD3
|
sterile alpha motif domain containing 3 |
| chr1_+_240255166 | 0.32 |
ENST00000319653.9
|
FMN2
|
formin 2 |
| chr11_-_76381029 | 0.32 |
ENST00000407242.2
ENST00000421973.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr5_-_173043591 | 0.31 |
ENST00000285908.5
ENST00000480951.1 ENST00000311086.4 |
BOD1
|
biorientation of chromosomes in cell division 1 |
| chr19_+_55385682 | 0.31 |
ENST00000391726.3
|
FCAR
|
Fc fragment of IgA, receptor for |
| chr19_+_49588677 | 0.30 |
ENST00000598984.1
ENST00000598441.1 |
SNRNP70
|
small nuclear ribonucleoprotein 70kDa (U1) |
| chr1_-_86848760 | 0.29 |
ENST00000460698.2
|
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chr6_-_71666732 | 0.29 |
ENST00000230053.6
|
B3GAT2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr3_+_178276488 | 0.29 |
ENST00000432997.1
ENST00000455865.1 |
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr11_-_76381781 | 0.28 |
ENST00000260061.5
ENST00000404995.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr12_-_10607084 | 0.27 |
ENST00000408006.3
ENST00000544822.1 ENST00000536188.1 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr11_+_62009653 | 0.26 |
ENST00000244926.3
|
SCGB1D2
|
secretoglobin, family 1D, member 2 |
| chr12_+_93096619 | 0.26 |
ENST00000397833.3
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr10_-_75401500 | 0.23 |
ENST00000359322.4
|
MYOZ1
|
myozenin 1 |
| chr1_+_152957707 | 0.23 |
ENST00000368762.1
|
SPRR1A
|
small proline-rich protein 1A |
| chr6_+_28249299 | 0.22 |
ENST00000405948.2
|
PGBD1
|
piggyBac transposable element derived 1 |
| chr8_-_93029520 | 0.22 |
ENST00000521553.1
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr5_+_140743859 | 0.22 |
ENST00000518069.1
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr11_-_5255696 | 0.20 |
ENST00000292901.3
ENST00000417377.1 |
HBD
|
hemoglobin, delta |
| chr12_+_57810198 | 0.20 |
ENST00000598001.1
|
AC126614.1
|
HCG1818482; Uncharacterized protein |
| chrX_+_129040122 | 0.20 |
ENST00000394422.3
ENST00000371051.5 |
UTP14A
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
| chr22_-_30662828 | 0.19 |
ENST00000403463.1
ENST00000215781.2 |
OSM
|
oncostatin M |
| chr19_+_49588690 | 0.18 |
ENST00000221448.5
|
SNRNP70
|
small nuclear ribonucleoprotein 70kDa (U1) |
| chr14_+_22356029 | 0.18 |
ENST00000390437.2
|
TRAV12-2
|
T cell receptor alpha variable 12-2 |
| chr6_+_28249332 | 0.17 |
ENST00000259883.3
|
PGBD1
|
piggyBac transposable element derived 1 |
| chr9_-_123638633 | 0.17 |
ENST00000456291.1
|
PHF19
|
PHD finger protein 19 |
| chr11_-_26593677 | 0.17 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr15_+_76196234 | 0.16 |
ENST00000540507.1
ENST00000565036.1 ENST00000569054.1 |
FBXO22
|
F-box protein 22 |
| chrX_+_129040094 | 0.16 |
ENST00000425117.2
|
UTP14A
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
| chr14_+_62462541 | 0.16 |
ENST00000430451.2
|
SYT16
|
synaptotagmin XVI |
| chr22_-_17302589 | 0.16 |
ENST00000331428.5
|
XKR3
|
XK, Kell blood group complex subunit-related family, member 3 |
| chr9_-_95244781 | 0.15 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr1_-_200379129 | 0.15 |
ENST00000367353.1
|
ZNF281
|
zinc finger protein 281 |
| chr14_+_21249200 | 0.15 |
ENST00000304677.2
|
RNASE6
|
ribonuclease, RNase A family, k6 |
| chr7_+_142457315 | 0.15 |
ENST00000486171.1
ENST00000311737.7 |
PRSS1
|
protease, serine, 1 (trypsin 1) |
| chr1_+_12806141 | 0.15 |
ENST00000288048.5
|
C1orf158
|
chromosome 1 open reading frame 158 |
| chr1_-_200379104 | 0.15 |
ENST00000367352.3
|
ZNF281
|
zinc finger protein 281 |
| chr3_+_136676707 | 0.15 |
ENST00000329582.4
|
IL20RB
|
interleukin 20 receptor beta |
| chr12_-_110434096 | 0.14 |
ENST00000320063.9
ENST00000457474.2 ENST00000547815.1 ENST00000361006.5 |
GIT2
|
G protein-coupled receptor kinase interacting ArfGAP 2 |
| chr12_+_55248289 | 0.14 |
ENST00000308796.6
|
MUCL1
|
mucin-like 1 |
| chrX_-_115594160 | 0.14 |
ENST00000371894.4
|
CXorf61
|
cancer/testis antigen 83 |
| chr16_+_12058961 | 0.13 |
ENST00000053243.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr3_-_185655795 | 0.13 |
ENST00000342294.4
ENST00000382191.4 ENST00000453386.2 |
TRA2B
|
transformer 2 beta homolog (Drosophila) |
| chr16_+_12059050 | 0.13 |
ENST00000396495.3
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr3_+_136676851 | 0.13 |
ENST00000309741.5
|
IL20RB
|
interleukin 20 receptor beta |
| chr3_+_174577070 | 0.12 |
ENST00000454872.1
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2 |
| chr4_-_142134031 | 0.12 |
ENST00000420921.2
|
RNF150
|
ring finger protein 150 |
| chr12_-_10605929 | 0.12 |
ENST00000347831.5
ENST00000359151.3 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr1_-_212004090 | 0.12 |
ENST00000366997.4
|
LPGAT1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr2_+_171034646 | 0.12 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
| chr11_-_111649015 | 0.12 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr2_+_102615416 | 0.12 |
ENST00000393414.2
|
IL1R2
|
interleukin 1 receptor, type II |
| chr12_-_11150474 | 0.11 |
ENST00000538986.1
|
TAS2R20
|
taste receptor, type 2, member 20 |
| chr4_-_74486217 | 0.11 |
ENST00000335049.5
ENST00000307439.5 |
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr6_-_15548591 | 0.11 |
ENST00000509674.1
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr11_-_26593649 | 0.09 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr2_+_171036635 | 0.09 |
ENST00000484338.2
ENST00000334231.6 |
MYO3B
|
myosin IIIB |
| chr17_-_15168624 | 0.08 |
ENST00000312280.3
ENST00000494511.1 ENST00000580584.1 |
PMP22
|
peripheral myelin protein 22 |
| chr1_-_112106556 | 0.08 |
ENST00000443498.1
|
ADORA3
|
adenosine A3 receptor |
| chr7_-_105029812 | 0.07 |
ENST00000482897.1
|
SRPK2
|
SRSF protein kinase 2 |
| chr6_-_46048116 | 0.07 |
ENST00000185206.6
|
CLIC5
|
chloride intracellular channel 5 |
| chr20_-_49575058 | 0.07 |
ENST00000371584.4
ENST00000371583.5 ENST00000413082.1 |
DPM1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr3_-_33686925 | 0.06 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr7_-_92848858 | 0.06 |
ENST00000440868.1
|
HEPACAM2
|
HEPACAM family member 2 |
| chr7_-_92848878 | 0.06 |
ENST00000341723.4
|
HEPACAM2
|
HEPACAM family member 2 |
| chr10_+_78078088 | 0.06 |
ENST00000496424.2
|
C10orf11
|
chromosome 10 open reading frame 11 |
| chr11_+_125496619 | 0.05 |
ENST00000532669.1
ENST00000278916.3 |
CHEK1
|
checkpoint kinase 1 |
| chr11_-_18062872 | 0.04 |
ENST00000250018.2
|
TPH1
|
tryptophan hydroxylase 1 |
| chr20_-_49575081 | 0.04 |
ENST00000371588.5
ENST00000371582.4 |
DPM1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr11_-_5255861 | 0.04 |
ENST00000380299.3
|
HBD
|
hemoglobin, delta |
| chr11_+_125496124 | 0.04 |
ENST00000533778.2
ENST00000534070.1 |
CHEK1
|
checkpoint kinase 1 |
| chr9_-_114090713 | 0.04 |
ENST00000302681.1
ENST00000374428.1 |
OR2K2
|
olfactory receptor, family 2, subfamily K, member 2 |
| chr17_-_38821373 | 0.02 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr20_-_48747662 | 0.02 |
ENST00000371656.2
|
TMEM189
|
transmembrane protein 189 |
| chr1_+_202789394 | 0.02 |
ENST00000330493.5
|
RP11-480I12.4
|
Putative inactive alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase-like protein LOC641515 |
| chr16_-_11876408 | 0.01 |
ENST00000396516.2
|
ZC3H7A
|
zinc finger CCCH-type containing 7A |
| chr1_+_152975488 | 0.01 |
ENST00000542696.1
|
SPRR3
|
small proline-rich protein 3 |
| chr4_-_74486109 | 0.01 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr4_-_74486347 | 0.00 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr6_+_26183958 | 0.00 |
ENST00000356530.3
|
HIST1H2BE
|
histone cluster 1, H2be |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0070340 | central nervous system myelin formation(GO:0032289) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.7 | 2.7 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.3 | 3.0 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.3 | 1.1 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.3 | 1.1 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.1 | 0.4 | GO:0018307 | tRNA wobble position uridine thiolation(GO:0002143) enzyme active site formation(GO:0018307) |
| 0.1 | 0.4 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.1 | 0.9 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.1 | 0.3 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.1 | 0.5 | GO:0061083 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.1 | 0.7 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 0.5 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.1 | 0.3 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.1 | 0.5 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 | 0.9 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 0.4 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 0.6 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.5 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 1.2 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.5 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.8 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 1.0 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.6 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.2 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.8 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 1.1 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.0 | 1.1 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.4 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 1.3 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.9 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 | 1.9 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.2 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.7 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.0 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.0 | 0.3 | GO:0002385 | mucosal immune response(GO:0002385) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.1 | 1.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 1.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.6 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 2.6 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.5 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.5 | GO:0002080 | acrosomal membrane(GO:0002080) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.0 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.4 | 3.5 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.3 | 0.8 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 0.7 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.1 | 1.3 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.4 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 1.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.4 | GO:0016295 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.1 | 0.3 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.5 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 0.8 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.4 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.3 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.9 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 0.4 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 3.8 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 0.7 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.1 | 1.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.5 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.6 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 2.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 1.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 0.9 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 1.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 1.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.5 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 1.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.6 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.8 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |