Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
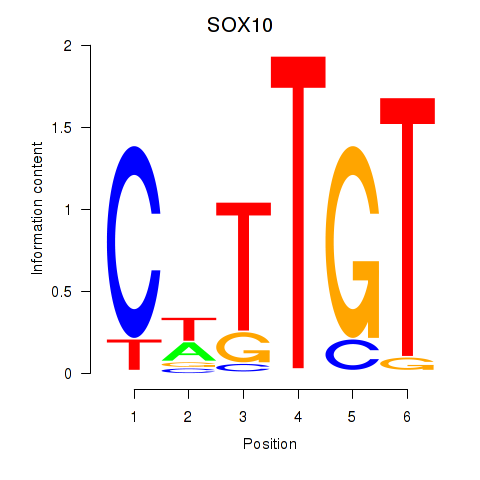
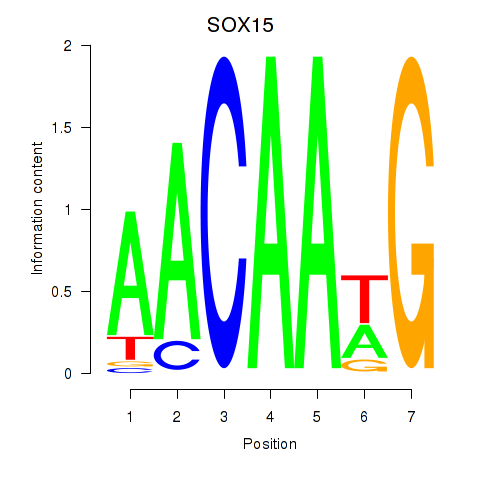
Results for SOX10_SOX15
Z-value: 0.56
Transcription factors associated with SOX10_SOX15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX10
|
ENSG00000100146.12 | SRY-box transcription factor 10 |
|
SOX15
|
ENSG00000129194.3 | SRY-box transcription factor 15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX15 | hg19_v2_chr17_-_7493390_7493488 | -0.46 | 2.0e-02 | Click! |
| SOX10 | hg19_v2_chr22_-_38380543_38380569 | -0.37 | 7.3e-02 | Click! |
Activity profile of SOX10_SOX15 motif
Sorted Z-values of SOX10_SOX15 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_188312971 | 3.49 |
ENST00000410068.1
ENST00000447403.1 ENST00000410102.1 |
CALCRL
|
calcitonin receptor-like |
| chr7_-_27219849 | 2.39 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr4_+_41614909 | 1.99 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr8_-_124553437 | 1.97 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr4_+_41614720 | 1.90 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr20_-_45985172 | 1.76 |
ENST00000536340.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr15_-_52944231 | 1.72 |
ENST00000546305.2
|
FAM214A
|
family with sequence similarity 214, member A |
| chr4_-_186877502 | 1.56 |
ENST00000431902.1
ENST00000284776.7 ENST00000415274.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr8_-_80993010 | 1.49 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr20_-_39317868 | 1.28 |
ENST00000373313.2
|
MAFB
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B |
| chr20_-_45984401 | 1.23 |
ENST00000311275.7
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr3_-_18466026 | 1.08 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr12_+_53443680 | 1.00 |
ENST00000314250.6
ENST00000451358.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr12_+_53443963 | 0.98 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr4_-_186732048 | 0.96 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr18_+_6729698 | 0.91 |
ENST00000383472.4
|
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr11_-_66445219 | 0.89 |
ENST00000525754.1
ENST00000531969.1 ENST00000524637.1 ENST00000531036.2 ENST00000310046.4 |
RBM4B
|
RNA binding motif protein 4B |
| chr15_+_57210961 | 0.87 |
ENST00000557843.1
|
TCF12
|
transcription factor 12 |
| chr6_+_122720681 | 0.84 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr11_-_130184555 | 0.84 |
ENST00000525842.1
|
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr1_+_33722080 | 0.82 |
ENST00000483388.1
ENST00000539719.1 |
ZNF362
|
zinc finger protein 362 |
| chr9_-_14308004 | 0.73 |
ENST00000493697.1
|
NFIB
|
nuclear factor I/B |
| chr11_-_130184470 | 0.66 |
ENST00000357899.4
ENST00000397753.1 |
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr3_+_151986709 | 0.63 |
ENST00000495875.2
ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr14_+_54976603 | 0.63 |
ENST00000557317.1
|
CGRRF1
|
cell growth regulator with ring finger domain 1 |
| chr18_-_22932080 | 0.61 |
ENST00000584787.1
ENST00000361524.3 ENST00000538137.2 |
ZNF521
|
zinc finger protein 521 |
| chr15_-_56035177 | 0.61 |
ENST00000389286.4
ENST00000561292.1 |
PRTG
|
protogenin |
| chr2_+_70056762 | 0.60 |
ENST00000282570.3
|
GMCL1
|
germ cell-less, spermatogenesis associated 1 |
| chr15_-_52043722 | 0.57 |
ENST00000454181.2
|
LYSMD2
|
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr10_-_97321165 | 0.56 |
ENST00000306402.6
|
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr1_+_185703513 | 0.56 |
ENST00000271588.4
ENST00000367492.2 |
HMCN1
|
hemicentin 1 |
| chrX_+_12993202 | 0.55 |
ENST00000451311.2
ENST00000380636.1 |
TMSB4X
|
thymosin beta 4, X-linked |
| chr14_+_77228532 | 0.52 |
ENST00000167106.4
ENST00000554237.1 |
VASH1
|
vasohibin 1 |
| chr4_-_138453606 | 0.51 |
ENST00000412923.2
ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18
|
protocadherin 18 |
| chr7_+_102553430 | 0.51 |
ENST00000339431.4
ENST00000249377.4 |
LRRC17
|
leucine rich repeat containing 17 |
| chr11_-_89224488 | 0.49 |
ENST00000534731.1
ENST00000527626.1 |
NOX4
|
NADPH oxidase 4 |
| chr11_-_89224299 | 0.46 |
ENST00000343727.5
ENST00000531342.1 ENST00000375979.3 |
NOX4
|
NADPH oxidase 4 |
| chr6_+_119215308 | 0.46 |
ENST00000229595.5
|
ASF1A
|
anti-silencing function 1A histone chaperone |
| chr3_+_141105235 | 0.46 |
ENST00000503809.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr19_+_1249869 | 0.43 |
ENST00000591446.2
|
MIDN
|
midnolin |
| chr2_+_32288657 | 0.43 |
ENST00000345662.1
|
SPAST
|
spastin |
| chr5_-_111092930 | 0.42 |
ENST00000257435.7
|
NREP
|
neuronal regeneration related protein |
| chr15_+_57210818 | 0.42 |
ENST00000438423.2
ENST00000267811.5 ENST00000452095.2 ENST00000559609.1 ENST00000333725.5 |
TCF12
|
transcription factor 12 |
| chr5_-_111093167 | 0.42 |
ENST00000446294.2
ENST00000419114.2 |
NREP
|
neuronal regeneration related protein |
| chr4_+_95129061 | 0.41 |
ENST00000354268.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr4_+_95128996 | 0.41 |
ENST00000457823.2
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr5_-_111093081 | 0.41 |
ENST00000453526.2
ENST00000509427.1 |
NREP
|
neuronal regeneration related protein |
| chr13_+_37393351 | 0.40 |
ENST00000255476.2
|
RFXAP
|
regulatory factor X-associated protein |
| chr2_+_66662510 | 0.40 |
ENST00000272369.9
ENST00000407092.2 |
MEIS1
|
Meis homeobox 1 |
| chr2_+_66662690 | 0.40 |
ENST00000488550.1
|
MEIS1
|
Meis homeobox 1 |
| chr18_+_6729725 | 0.39 |
ENST00000400091.2
ENST00000583410.1 ENST00000584387.1 |
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr2_+_120517174 | 0.39 |
ENST00000263708.2
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr8_+_21777159 | 0.39 |
ENST00000434536.1
ENST00000252512.9 |
XPO7
|
exportin 7 |
| chr2_+_69240302 | 0.39 |
ENST00000303714.4
|
ANTXR1
|
anthrax toxin receptor 1 |
| chrX_-_32173579 | 0.38 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr4_+_55524085 | 0.38 |
ENST00000412167.2
ENST00000288135.5 |
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
| chr1_+_163038565 | 0.38 |
ENST00000421743.2
|
RGS4
|
regulator of G-protein signaling 4 |
| chr5_-_111092873 | 0.38 |
ENST00000509025.1
ENST00000515855.1 |
NREP
|
neuronal regeneration related protein |
| chr22_+_39052632 | 0.38 |
ENST00000411557.1
ENST00000396811.2 ENST00000216029.3 ENST00000416285.1 |
CBY1
|
chibby homolog 1 (Drosophila) |
| chr7_+_116312411 | 0.38 |
ENST00000456159.1
ENST00000397752.3 ENST00000318493.6 |
MET
|
met proto-oncogene |
| chr10_-_99393208 | 0.37 |
ENST00000307450.6
|
MORN4
|
MORN repeat containing 4 |
| chr1_+_93913713 | 0.37 |
ENST00000604705.1
ENST00000370253.2 |
FNBP1L
|
formin binding protein 1-like |
| chr12_+_32260085 | 0.37 |
ENST00000548411.1
ENST00000281474.5 ENST00000551086.1 |
BICD1
|
bicaudal D homolog 1 (Drosophila) |
| chr11_-_89224508 | 0.37 |
ENST00000525196.1
|
NOX4
|
NADPH oxidase 4 |
| chr2_+_32288725 | 0.36 |
ENST00000315285.3
|
SPAST
|
spastin |
| chrX_+_12993336 | 0.36 |
ENST00000380635.1
|
TMSB4X
|
thymosin beta 4, X-linked |
| chr8_-_17579726 | 0.36 |
ENST00000381861.3
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr4_+_144303093 | 0.36 |
ENST00000505913.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chrX_+_107069063 | 0.35 |
ENST00000262843.6
|
MID2
|
midline 2 |
| chrX_-_128788914 | 0.35 |
ENST00000429967.1
ENST00000307484.6 |
APLN
|
apelin |
| chr3_+_107241882 | 0.34 |
ENST00000416476.2
|
BBX
|
bobby sox homolog (Drosophila) |
| chr10_+_99079008 | 0.34 |
ENST00000371021.3
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas |
| chr11_-_89224638 | 0.34 |
ENST00000535633.1
ENST00000263317.4 |
NOX4
|
NADPH oxidase 4 |
| chrX_+_80457442 | 0.33 |
ENST00000373212.5
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like |
| chrX_-_24665208 | 0.33 |
ENST00000356768.4
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr2_-_183387430 | 0.32 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr3_-_141747950 | 0.32 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr4_+_71588372 | 0.32 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr2_+_181845843 | 0.32 |
ENST00000602710.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr13_+_78315295 | 0.31 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr4_+_95128748 | 0.31 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr17_-_46690839 | 0.30 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr10_+_35484793 | 0.30 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr12_-_12419703 | 0.29 |
ENST00000543091.1
ENST00000261349.4 |
LRP6
|
low density lipoprotein receptor-related protein 6 |
| chr3_-_167813672 | 0.29 |
ENST00000470487.1
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr1_-_26233423 | 0.29 |
ENST00000357865.2
|
STMN1
|
stathmin 1 |
| chr7_-_140178775 | 0.29 |
ENST00000474576.1
ENST00000473444.1 ENST00000471104.1 |
MKRN1
|
makorin ring finger protein 1 |
| chr6_+_56911476 | 0.29 |
ENST00000545356.1
|
KIAA1586
|
KIAA1586 |
| chr15_+_52043758 | 0.29 |
ENST00000249700.4
ENST00000539962.2 |
TMOD2
|
tropomodulin 2 (neuronal) |
| chr11_-_94965667 | 0.28 |
ENST00000542176.1
ENST00000278499.2 |
SESN3
|
sestrin 3 |
| chr11_+_36317830 | 0.28 |
ENST00000530639.1
|
PRR5L
|
proline rich 5 like |
| chr3_+_113616317 | 0.28 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr7_-_5821225 | 0.28 |
ENST00000416985.1
|
RNF216
|
ring finger protein 216 |
| chr2_-_165424973 | 0.28 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr14_-_22005018 | 0.28 |
ENST00000546363.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr18_+_72201664 | 0.28 |
ENST00000358821.3
|
CNDP1
|
carnosine dipeptidase 1 (metallopeptidase M20 family) |
| chr6_-_111888474 | 0.27 |
ENST00000368735.1
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr10_+_22605374 | 0.27 |
ENST00000448361.1
|
COMMD3
|
COMM domain containing 3 |
| chr20_-_23066953 | 0.27 |
ENST00000246006.4
|
CD93
|
CD93 molecule |
| chrX_-_10645773 | 0.27 |
ENST00000453318.2
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr10_+_35415719 | 0.27 |
ENST00000474362.1
ENST00000374721.3 |
CREM
|
cAMP responsive element modulator |
| chr10_+_22605304 | 0.26 |
ENST00000475460.2
ENST00000602390.1 ENST00000489125.2 ENST00000456711.1 ENST00000444869.1 |
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 readthrough COMM domain containing 3 |
| chr6_+_83777374 | 0.26 |
ENST00000349129.2
ENST00000237163.5 ENST00000536812.1 |
DOPEY1
|
dopey family member 1 |
| chr16_-_15736881 | 0.26 |
ENST00000540441.2
|
KIAA0430
|
KIAA0430 |
| chr7_-_121944491 | 0.26 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr4_+_90033968 | 0.25 |
ENST00000317005.2
|
TIGD2
|
tigger transposable element derived 2 |
| chr6_+_30848557 | 0.25 |
ENST00000460944.2
ENST00000324771.8 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr6_-_41863098 | 0.25 |
ENST00000373006.1
|
USP49
|
ubiquitin specific peptidase 49 |
| chrX_-_21776281 | 0.25 |
ENST00000379494.3
|
SMPX
|
small muscle protein, X-linked |
| chr2_-_70475701 | 0.25 |
ENST00000282574.4
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr3_+_107241783 | 0.25 |
ENST00000415149.2
ENST00000402543.1 ENST00000325805.8 ENST00000427402.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr7_-_99679324 | 0.24 |
ENST00000292393.5
ENST00000413658.2 ENST00000412947.1 ENST00000441298.1 ENST00000449785.1 ENST00000299667.4 ENST00000424697.1 |
ZNF3
|
zinc finger protein 3 |
| chr6_+_74405501 | 0.24 |
ENST00000437994.2
ENST00000422508.2 |
CD109
|
CD109 molecule |
| chr17_-_39471947 | 0.24 |
ENST00000334202.3
|
KRTAP17-1
|
keratin associated protein 17-1 |
| chr10_+_35415978 | 0.23 |
ENST00000429130.3
ENST00000469949.2 ENST00000460270.1 |
CREM
|
cAMP responsive element modulator |
| chr17_+_67498538 | 0.23 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr9_-_35115836 | 0.23 |
ENST00000378566.1
ENST00000378554.2 ENST00000322813.5 |
FAM214B
|
family with sequence similarity 214, member B |
| chrX_-_63425561 | 0.23 |
ENST00000374869.3
ENST00000330258.3 |
AMER1
|
APC membrane recruitment protein 1 |
| chr14_-_71107921 | 0.23 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr6_-_112575912 | 0.23 |
ENST00000522006.1
ENST00000230538.7 ENST00000519932.1 |
LAMA4
|
laminin, alpha 4 |
| chr14_-_106066376 | 0.23 |
ENST00000412518.1
ENST00000428654.1 ENST00000427543.1 ENST00000420153.1 ENST00000577108.1 ENST00000576077.1 |
AL928742.12
IGHE
|
AL928742.12 immunoglobulin heavy constant epsilon |
| chr5_+_43602750 | 0.23 |
ENST00000505678.2
ENST00000512422.1 ENST00000264663.5 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr13_+_78315466 | 0.23 |
ENST00000314070.5
ENST00000462234.1 |
SLAIN1
|
SLAIN motif family, member 1 |
| chr10_-_128359074 | 0.23 |
ENST00000544758.1
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr20_+_11898507 | 0.23 |
ENST00000378226.2
|
BTBD3
|
BTB (POZ) domain containing 3 |
| chr17_+_19030782 | 0.23 |
ENST00000344415.4
ENST00000577213.1 |
GRAPL
|
GRB2-related adaptor protein-like |
| chr16_-_4852915 | 0.22 |
ENST00000322048.7
|
ROGDI
|
rogdi homolog (Drosophila) |
| chr6_+_74405804 | 0.22 |
ENST00000287097.5
|
CD109
|
CD109 molecule |
| chr10_-_14646388 | 0.22 |
ENST00000468747.1
ENST00000378467.4 |
FAM107B
|
family with sequence similarity 107, member B |
| chr15_+_84115868 | 0.21 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr19_+_34287751 | 0.21 |
ENST00000590771.1
ENST00000589786.1 ENST00000284006.6 ENST00000588881.1 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chrX_-_106960285 | 0.21 |
ENST00000503515.1
ENST00000372397.2 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr16_+_53242350 | 0.21 |
ENST00000565442.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr16_+_30406721 | 0.21 |
ENST00000320159.2
|
ZNF48
|
zinc finger protein 48 |
| chr13_-_33780133 | 0.21 |
ENST00000399365.3
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr7_-_15601595 | 0.20 |
ENST00000342526.3
|
AGMO
|
alkylglycerol monooxygenase |
| chrX_+_129473859 | 0.20 |
ENST00000424447.1
|
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr1_+_66797687 | 0.20 |
ENST00000371045.5
ENST00000531025.1 ENST00000526197.1 |
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr11_+_63706444 | 0.20 |
ENST00000377793.4
ENST00000456907.2 ENST00000539656.1 |
NAA40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr3_-_129407535 | 0.20 |
ENST00000432054.2
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr17_+_73452545 | 0.20 |
ENST00000314256.7
|
KIAA0195
|
KIAA0195 |
| chr18_+_72201829 | 0.20 |
ENST00000582365.1
|
CNDP1
|
carnosine dipeptidase 1 (metallopeptidase M20 family) |
| chr4_+_106067943 | 0.20 |
ENST00000380013.4
ENST00000394764.1 ENST00000413648.2 |
TET2
|
tet methylcytosine dioxygenase 2 |
| chr12_-_90049878 | 0.20 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr3_+_107096188 | 0.19 |
ENST00000261058.1
|
CCDC54
|
coiled-coil domain containing 54 |
| chr13_+_30002846 | 0.19 |
ENST00000542829.1
|
MTUS2
|
microtubule associated tumor suppressor candidate 2 |
| chr7_+_116660246 | 0.19 |
ENST00000434836.1
ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr1_-_147245484 | 0.19 |
ENST00000271348.2
|
GJA5
|
gap junction protein, alpha 5, 40kDa |
| chr5_+_78532003 | 0.19 |
ENST00000396137.4
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr17_+_75447326 | 0.18 |
ENST00000591088.1
|
SEPT9
|
septin 9 |
| chr4_+_118955500 | 0.18 |
ENST00000296499.5
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr12_-_90049828 | 0.18 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr5_+_147774275 | 0.18 |
ENST00000513826.1
|
FBXO38
|
F-box protein 38 |
| chr2_-_70475730 | 0.18 |
ENST00000445587.1
ENST00000433529.2 ENST00000415783.2 |
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr2_+_177015950 | 0.18 |
ENST00000306324.3
|
HOXD4
|
homeobox D4 |
| chr18_-_74728998 | 0.18 |
ENST00000359645.3
ENST00000397875.3 ENST00000397869.3 ENST00000578193.1 ENST00000578873.1 ENST00000397866.4 ENST00000528160.1 ENST00000527041.1 ENST00000526111.1 ENST00000397865.5 ENST00000382582.3 |
MBP
|
myelin basic protein |
| chr3_+_189507460 | 0.18 |
ENST00000434928.1
|
TP63
|
tumor protein p63 |
| chrX_-_24665353 | 0.18 |
ENST00000379144.2
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr3_+_14860469 | 0.18 |
ENST00000285046.5
|
FGD5
|
FYVE, RhoGEF and PH domain containing 5 |
| chr22_-_41636892 | 0.18 |
ENST00000455425.1
|
CHADL
|
chondroadherin-like |
| chr2_+_198669365 | 0.18 |
ENST00000428675.1
|
PLCL1
|
phospholipase C-like 1 |
| chr16_-_28303360 | 0.17 |
ENST00000501520.1
|
RP11-57A19.2
|
RP11-57A19.2 |
| chr17_-_46623441 | 0.17 |
ENST00000330070.4
|
HOXB2
|
homeobox B2 |
| chr12_-_71003568 | 0.17 |
ENST00000547715.1
ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr17_+_36584662 | 0.17 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr22_+_19710468 | 0.17 |
ENST00000366425.3
|
GP1BB
|
glycoprotein Ib (platelet), beta polypeptide |
| chr18_-_31802282 | 0.17 |
ENST00000535475.1
|
NOL4
|
nucleolar protein 4 |
| chr17_-_18950310 | 0.17 |
ENST00000573099.1
|
GRAP
|
GRB2-related adaptor protein |
| chr21_+_37442239 | 0.17 |
ENST00000530908.1
ENST00000290349.6 ENST00000439427.2 ENST00000399191.3 |
CBR1
|
carbonyl reductase 1 |
| chr4_+_113152978 | 0.17 |
ENST00000309703.6
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr3_+_183353356 | 0.16 |
ENST00000242810.6
ENST00000493074.1 ENST00000437402.1 ENST00000454495.2 ENST00000473045.1 ENST00000468101.1 ENST00000427201.2 ENST00000482138.1 ENST00000454652.2 |
KLHL24
|
kelch-like family member 24 |
| chr13_+_49684445 | 0.16 |
ENST00000398316.3
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr14_+_100070869 | 0.16 |
ENST00000502101.2
|
RP11-543C4.1
|
RP11-543C4.1 |
| chr12_-_102874330 | 0.16 |
ENST00000307046.8
|
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr4_+_113218497 | 0.16 |
ENST00000458497.1
ENST00000177648.9 ENST00000504176.2 |
ALPK1
|
alpha-kinase 1 |
| chr6_-_112575838 | 0.16 |
ENST00000455073.1
|
LAMA4
|
laminin, alpha 4 |
| chr2_-_86564776 | 0.16 |
ENST00000165698.5
ENST00000541910.1 ENST00000535845.1 |
REEP1
|
receptor accessory protein 1 |
| chrX_+_37639264 | 0.16 |
ENST00000378588.4
|
CYBB
|
cytochrome b-245, beta polypeptide |
| chr19_+_50706866 | 0.16 |
ENST00000440075.2
ENST00000376970.2 ENST00000425460.1 ENST00000599920.1 ENST00000601313.1 |
MYH14
|
myosin, heavy chain 14, non-muscle |
| chrX_+_10124977 | 0.16 |
ENST00000380833.4
|
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr4_+_150999418 | 0.15 |
ENST00000296550.7
|
DCLK2
|
doublecortin-like kinase 2 |
| chr18_+_3448455 | 0.15 |
ENST00000549780.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr15_-_82338460 | 0.15 |
ENST00000558133.1
ENST00000329713.4 |
MEX3B
|
mex-3 RNA binding family member B |
| chr4_-_170192185 | 0.15 |
ENST00000284637.9
|
SH3RF1
|
SH3 domain containing ring finger 1 |
| chrY_+_15815447 | 0.15 |
ENST00000284856.3
|
TMSB4Y
|
thymosin beta 4, Y-linked |
| chr7_-_111846435 | 0.15 |
ENST00000437633.1
ENST00000428084.1 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr4_+_113152881 | 0.15 |
ENST00000274000.5
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr1_-_53793584 | 0.15 |
ENST00000354412.3
ENST00000347547.2 ENST00000306052.6 |
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr17_+_73452695 | 0.15 |
ENST00000582186.1
ENST00000582455.1 ENST00000581252.1 ENST00000579208.1 |
KIAA0195
|
KIAA0195 |
| chr8_+_107738240 | 0.15 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chrX_+_129473916 | 0.15 |
ENST00000545805.1
ENST00000543953.1 ENST00000218197.5 |
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr1_+_228337553 | 0.15 |
ENST00000366714.2
|
GJC2
|
gap junction protein, gamma 2, 47kDa |
| chr14_-_67981870 | 0.15 |
ENST00000555994.1
|
TMEM229B
|
transmembrane protein 229B |
| chrX_-_117107542 | 0.15 |
ENST00000371878.1
|
KLHL13
|
kelch-like family member 13 |
| chr15_+_77224045 | 0.15 |
ENST00000320963.5
ENST00000394883.3 |
RCN2
|
reticulocalbin 2, EF-hand calcium binding domain |
| chr1_+_212797789 | 0.15 |
ENST00000294829.3
|
FAM71A
|
family with sequence similarity 71, member A |
| chr1_+_93645314 | 0.15 |
ENST00000343253.7
|
CCDC18
|
coiled-coil domain containing 18 |
| chr17_+_75283973 | 0.15 |
ENST00000431235.2
ENST00000449803.2 |
SEPT9
|
septin 9 |
| chr14_-_23904861 | 0.15 |
ENST00000355349.3
|
MYH7
|
myosin, heavy chain 7, cardiac muscle, beta |
| chr1_+_66458072 | 0.15 |
ENST00000423207.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX10_SOX15
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.0 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.4 | 1.3 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.4 | 1.3 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.3 | 3.5 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.2 | 2.0 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.2 | 0.2 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.2 | 0.5 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 0.7 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 0.4 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.1 | 0.4 | GO:0036215 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.1 | 0.4 | GO:1900276 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.1 | 0.4 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.1 | 0.2 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 0.9 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.1 | 0.3 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.1 | 0.7 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.3 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.1 | 0.4 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.8 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.5 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 2.9 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.5 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 1.1 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.5 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 2.4 | GO:0060065 | uterus development(GO:0060065) |
| 0.1 | 0.2 | GO:0003193 | pulmonary valve formation(GO:0003193) regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) foramen ovale closure(GO:0035922) regulation of bundle of His cell action potential(GO:0098905) |
| 0.1 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.3 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 1.1 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.3 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.4 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 1.7 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.5 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.6 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.4 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.5 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.2 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.7 | GO:0060044 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.0 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.6 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:2001037 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.1 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.0 | 0.4 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.2 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.2 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.2 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.9 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.6 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.0 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.2 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.3 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.3 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.0 | 0.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.0 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.2 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.0 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.3 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.2 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.6 | GO:0050691 | regulation of defense response to virus by host(GO:0050691) |
| 0.0 | 0.2 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.2 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.0 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.0 | 0.0 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.4 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 3.2 | GO:0031032 | actomyosin structure organization(GO:0031032) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0039714 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.1 | 0.6 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.3 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 1.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.2 | GO:1990015 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.1 | 0.4 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 0.4 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.7 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.4 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 2.6 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.2 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 1.1 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 1.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.9 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.3 | GO:0031105 | septin complex(GO:0031105) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.5 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.2 | 0.6 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.4 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 1.7 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 0.5 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 2.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.8 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 0.3 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.1 | 3.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.4 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 1.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.2 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.0 | 0.2 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.2 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 0.2 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.0 | 0.3 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.0 | 0.1 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.4 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.4 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 1.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.4 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.9 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.5 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.7 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.8 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 1.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 3.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 1.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.4 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.5 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |