Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
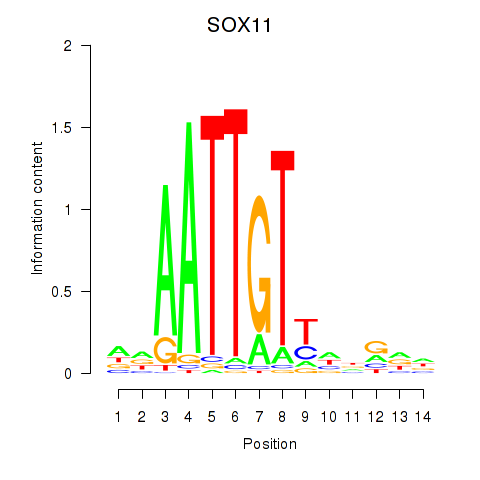
Results for SOX11
Z-value: 0.61
Transcription factors associated with SOX11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX11
|
ENSG00000176887.5 | SRY-box transcription factor 11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX11 | hg19_v2_chr2_+_5832799_5832799 | -0.30 | 1.5e-01 | Click! |
Activity profile of SOX11 motif
Sorted Z-values of SOX11 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_225266743 | 6.17 |
ENST00000409685.3
|
FAM124B
|
family with sequence similarity 124B |
| chr2_-_225266711 | 4.88 |
ENST00000389874.3
|
FAM124B
|
family with sequence similarity 124B |
| chr3_-_18466026 | 2.69 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr9_+_75766763 | 1.72 |
ENST00000456643.1
ENST00000415424.1 |
ANXA1
|
annexin A1 |
| chr4_-_90759440 | 1.34 |
ENST00000336904.3
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr11_+_92085262 | 1.22 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr12_+_32655048 | 1.12 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr17_-_66951474 | 1.09 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr6_-_76072719 | 1.07 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr9_+_71986182 | 0.79 |
ENST00000303068.7
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr12_+_60058458 | 0.79 |
ENST00000548610.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr4_-_110624564 | 0.79 |
ENST00000352981.3
ENST00000265164.2 ENST00000505486.1 |
CASP6
|
caspase 6, apoptosis-related cysteine peptidase |
| chr21_+_17792672 | 0.64 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr13_+_78315295 | 0.62 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr2_+_138721850 | 0.53 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chr5_+_53813536 | 0.52 |
ENST00000343017.6
ENST00000381410.4 ENST00000326277.3 |
SNX18
|
sorting nexin 18 |
| chr12_-_11287243 | 0.49 |
ENST00000539585.1
|
TAS2R30
|
taste receptor, type 2, member 30 |
| chr2_+_207804278 | 0.48 |
ENST00000272852.3
|
CPO
|
carboxypeptidase O |
| chr5_+_55149150 | 0.47 |
ENST00000297015.3
|
IL31RA
|
interleukin 31 receptor A |
| chr13_+_78315466 | 0.47 |
ENST00000314070.5
ENST00000462234.1 |
SLAIN1
|
SLAIN motif family, member 1 |
| chr5_+_140213815 | 0.41 |
ENST00000525929.1
ENST00000378125.3 |
PCDHA7
|
protocadherin alpha 7 |
| chr11_+_7110165 | 0.41 |
ENST00000306904.5
|
RBMXL2
|
RNA binding motif protein, X-linked-like 2 |
| chrX_+_102192200 | 0.39 |
ENST00000218249.5
|
RAB40AL
|
RAB40A, member RAS oncogene family-like |
| chr13_-_103019744 | 0.38 |
ENST00000437115.2
|
FGF14-IT1
|
FGF14 intronic transcript 1 (non-protein coding) |
| chr5_+_140248518 | 0.38 |
ENST00000398640.2
|
PCDHA11
|
protocadherin alpha 11 |
| chr15_+_49447947 | 0.38 |
ENST00000327171.3
ENST00000560654.1 |
GALK2
|
galactokinase 2 |
| chrX_-_100548045 | 0.38 |
ENST00000372907.3
ENST00000372905.2 |
TAF7L
|
TAF7-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 50kDa |
| chr1_+_145524891 | 0.37 |
ENST00000369304.3
|
ITGA10
|
integrin, alpha 10 |
| chr6_-_99873145 | 0.37 |
ENST00000369239.5
ENST00000438806.1 |
PNISR
|
PNN-interacting serine/arginine-rich protein |
| chr2_-_37068530 | 0.35 |
ENST00000593798.1
|
AC007382.1
|
Uncharacterized protein |
| chr17_+_38474489 | 0.35 |
ENST00000394089.2
ENST00000425707.3 |
RARA
|
retinoic acid receptor, alpha |
| chr2_+_138722028 | 0.34 |
ENST00000280096.5
|
HNMT
|
histamine N-methyltransferase |
| chr1_-_205744205 | 0.33 |
ENST00000446390.2
|
RAB7L1
|
RAB7, member RAS oncogene family-like 1 |
| chr10_+_35456444 | 0.33 |
ENST00000361599.4
|
CREM
|
cAMP responsive element modulator |
| chr5_-_54468974 | 0.33 |
ENST00000381375.2
ENST00000296733.1 ENST00000322374.6 ENST00000334206.5 ENST00000331730.3 |
CDC20B
|
cell division cycle 20B |
| chr8_+_104831554 | 0.33 |
ENST00000408894.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chrX_-_32173579 | 0.32 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr6_-_30585009 | 0.32 |
ENST00000376511.2
|
PPP1R10
|
protein phosphatase 1, regulatory subunit 10 |
| chr15_-_37392086 | 0.32 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chr12_+_28410128 | 0.32 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr17_+_47448102 | 0.32 |
ENST00000576461.1
|
RP11-81K2.1
|
Uncharacterized protein |
| chr14_+_21156915 | 0.31 |
ENST00000397990.4
ENST00000555597.1 |
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5 ribonuclease, RNase A family, 4 |
| chr11_+_22688150 | 0.31 |
ENST00000454584.2
|
GAS2
|
growth arrest-specific 2 |
| chr4_+_110749143 | 0.31 |
ENST00000317735.4
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr2_+_109237717 | 0.31 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr1_+_65730385 | 0.30 |
ENST00000263441.7
ENST00000395325.3 |
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr18_+_46065393 | 0.30 |
ENST00000256413.3
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr6_-_52705641 | 0.30 |
ENST00000370989.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr2_+_179318295 | 0.30 |
ENST00000442710.1
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr20_+_18794370 | 0.28 |
ENST00000377428.2
|
SCP2D1
|
SCP2 sterol-binding domain containing 1 |
| chr3_+_113616317 | 0.28 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr1_+_196621002 | 0.28 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr1_+_214776516 | 0.28 |
ENST00000366955.3
|
CENPF
|
centromere protein F, 350/400kDa |
| chr18_+_46065483 | 0.27 |
ENST00000382998.4
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr1_-_111061797 | 0.27 |
ENST00000369771.2
|
KCNA10
|
potassium voltage-gated channel, shaker-related subfamily, member 10 |
| chr4_+_130017268 | 0.26 |
ENST00000425929.1
ENST00000508673.1 ENST00000508622.1 |
C4orf33
|
chromosome 4 open reading frame 33 |
| chr1_-_205744574 | 0.26 |
ENST00000367139.3
ENST00000235932.4 ENST00000437324.2 ENST00000414729.1 |
RAB7L1
|
RAB7, member RAS oncogene family-like 1 |
| chr4_+_88754113 | 0.25 |
ENST00000560249.1
ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr1_-_24469602 | 0.25 |
ENST00000270800.1
|
IL22RA1
|
interleukin 22 receptor, alpha 1 |
| chr18_+_7754957 | 0.25 |
ENST00000400053.4
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr2_-_71222466 | 0.25 |
ENST00000606025.1
|
AC007040.11
|
Uncharacterized protein |
| chr8_-_54752406 | 0.24 |
ENST00000520188.1
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
| chr1_-_197169672 | 0.24 |
ENST00000367405.4
|
ZBTB41
|
zinc finger and BTB domain containing 41 |
| chr10_-_127505167 | 0.24 |
ENST00000368786.1
|
UROS
|
uroporphyrinogen III synthase |
| chr4_+_88754069 | 0.24 |
ENST00000395102.4
ENST00000497649.2 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr14_-_36789783 | 0.24 |
ENST00000605579.1
ENST00000604336.1 ENST00000359527.7 ENST00000603139.1 ENST00000318473.7 |
MBIP
|
MAP3K12 binding inhibitory protein 1 |
| chr4_-_170679024 | 0.23 |
ENST00000393381.2
|
C4orf27
|
chromosome 4 open reading frame 27 |
| chr14_-_36789865 | 0.23 |
ENST00000416007.4
|
MBIP
|
MAP3K12 binding inhibitory protein 1 |
| chr17_-_34344991 | 0.23 |
ENST00000591423.1
|
CCL23
|
chemokine (C-C motif) ligand 23 |
| chr2_+_102927962 | 0.22 |
ENST00000233954.1
ENST00000393393.3 ENST00000410040.1 |
IL1RL1
IL18R1
|
interleukin 1 receptor-like 1 interleukin 18 receptor 1 |
| chr3_-_133380731 | 0.22 |
ENST00000260810.5
|
TOPBP1
|
topoisomerase (DNA) II binding protein 1 |
| chr9_-_95166841 | 0.22 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chr1_-_241799232 | 0.20 |
ENST00000366553.1
|
CHML
|
choroideremia-like (Rab escort protein 2) |
| chr7_-_122635754 | 0.20 |
ENST00000249284.2
|
TAS2R16
|
taste receptor, type 2, member 16 |
| chr15_+_45028753 | 0.20 |
ENST00000338264.4
|
TRIM69
|
tripartite motif containing 69 |
| chr1_+_93645314 | 0.20 |
ENST00000343253.7
|
CCDC18
|
coiled-coil domain containing 18 |
| chr9_-_114521783 | 0.20 |
ENST00000394779.3
ENST00000394777.4 |
C9orf84
|
chromosome 9 open reading frame 84 |
| chr22_+_24407776 | 0.20 |
ENST00000405822.2
|
CABIN1
|
calcineurin binding protein 1 |
| chr13_-_30424821 | 0.19 |
ENST00000380680.4
|
UBL3
|
ubiquitin-like 3 |
| chr17_-_34345002 | 0.19 |
ENST00000293280.2
|
CCL23
|
chemokine (C-C motif) ligand 23 |
| chrX_+_27826107 | 0.17 |
ENST00000356790.2
|
MAGEB10
|
melanoma antigen family B, 10 |
| chr5_+_148521381 | 0.17 |
ENST00000504238.1
|
ABLIM3
|
actin binding LIM protein family, member 3 |
| chr3_+_101504200 | 0.17 |
ENST00000422132.1
|
NXPE3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr17_+_35732955 | 0.17 |
ENST00000300618.4
|
C17orf78
|
chromosome 17 open reading frame 78 |
| chr8_+_9413410 | 0.16 |
ENST00000520408.1
ENST00000310430.6 ENST00000522110.1 |
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr17_+_35732916 | 0.16 |
ENST00000586700.1
|
C17orf78
|
chromosome 17 open reading frame 78 |
| chr15_+_45028719 | 0.16 |
ENST00000560442.1
ENST00000558329.1 ENST00000561043.1 |
TRIM69
|
tripartite motif containing 69 |
| chr2_-_219157250 | 0.15 |
ENST00000434015.2
ENST00000444183.1 ENST00000420341.1 ENST00000453281.1 ENST00000258412.3 ENST00000440422.1 |
TMBIM1
|
transmembrane BAX inhibitor motif containing 1 |
| chr12_+_41221794 | 0.15 |
ENST00000547849.1
|
CNTN1
|
contactin 1 |
| chr1_+_196621156 | 0.15 |
ENST00000359637.2
|
CFH
|
complement factor H |
| chr2_-_20251744 | 0.15 |
ENST00000175091.4
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha |
| chr6_+_53883790 | 0.15 |
ENST00000509997.1
|
MLIP
|
muscular LMNA-interacting protein |
| chr18_+_22040620 | 0.15 |
ENST00000426880.2
|
HRH4
|
histamine receptor H4 |
| chr10_-_5446786 | 0.14 |
ENST00000479328.1
ENST00000380419.3 |
TUBAL3
|
tubulin, alpha-like 3 |
| chr15_-_64673665 | 0.14 |
ENST00000300035.4
|
KIAA0101
|
KIAA0101 |
| chrX_+_130192318 | 0.14 |
ENST00000370922.1
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr7_+_90012986 | 0.14 |
ENST00000416322.1
|
CLDN12
|
claudin 12 |
| chr1_+_198608146 | 0.14 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr15_-_78913628 | 0.14 |
ENST00000348639.3
|
CHRNA3
|
cholinergic receptor, nicotinic, alpha 3 (neuronal) |
| chr4_+_159443090 | 0.13 |
ENST00000343542.5
ENST00000470033.1 |
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chr12_-_122879969 | 0.12 |
ENST00000540304.1
|
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr10_+_5238793 | 0.11 |
ENST00000263126.1
|
AKR1C4
|
aldo-keto reductase family 1, member C4 |
| chr10_+_32873190 | 0.11 |
ENST00000375025.4
|
C10orf68
|
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
| chr11_-_64684672 | 0.11 |
ENST00000377264.3
ENST00000421419.2 |
ATG2A
|
autophagy related 2A |
| chr12_-_122240792 | 0.10 |
ENST00000545885.1
ENST00000542933.1 ENST00000428029.2 ENST00000541694.1 ENST00000536662.1 ENST00000535643.1 ENST00000541657.1 |
AC084018.1
RHOF
|
AC084018.1 ras homolog family member F (in filopodia) |
| chr5_+_140207536 | 0.10 |
ENST00000529310.1
ENST00000527624.1 |
PCDHA6
|
protocadherin alpha 6 |
| chrX_+_24483338 | 0.10 |
ENST00000379162.4
ENST00000441463.2 |
PDK3
|
pyruvate dehydrogenase kinase, isozyme 3 |
| chr9_+_27109133 | 0.10 |
ENST00000519097.1
ENST00000380036.4 |
TEK
|
TEK tyrosine kinase, endothelial |
| chr18_+_22040593 | 0.10 |
ENST00000256906.4
|
HRH4
|
histamine receptor H4 |
| chr5_-_86534822 | 0.09 |
ENST00000445770.2
|
AC008394.1
|
Uncharacterized protein |
| chrX_+_100645812 | 0.09 |
ENST00000427805.2
ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr6_+_53883708 | 0.09 |
ENST00000514921.1
ENST00000274897.5 ENST00000370877.2 |
MLIP
|
muscular LMNA-interacting protein |
| chrX_-_7895755 | 0.09 |
ENST00000444736.1
ENST00000537427.1 ENST00000442940.1 |
PNPLA4
|
patatin-like phospholipase domain containing 4 |
| chr7_-_81399355 | 0.09 |
ENST00000457544.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr6_-_46048116 | 0.09 |
ENST00000185206.6
|
CLIC5
|
chloride intracellular channel 5 |
| chr7_-_82792215 | 0.09 |
ENST00000333891.9
ENST00000423517.2 |
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr6_-_133055815 | 0.07 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr8_-_25281747 | 0.07 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr10_-_95242044 | 0.07 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr4_+_141294628 | 0.07 |
ENST00000512749.1
ENST00000608372.1 ENST00000506597.1 ENST00000394201.4 ENST00000510586.1 |
SCOC
|
short coiled-coil protein |
| chr2_+_47596287 | 0.07 |
ENST00000263735.4
|
EPCAM
|
epithelial cell adhesion molecule |
| chr14_+_39703112 | 0.06 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr3_-_49967292 | 0.06 |
ENST00000455683.2
|
MON1A
|
MON1 secretory trafficking family member A |
| chr4_+_159442878 | 0.06 |
ENST00000307765.5
ENST00000423548.1 |
RXFP1
|
relaxin/insulin-like family peptide receptor 1 |
| chr15_-_64673630 | 0.06 |
ENST00000558008.1
ENST00000559519.1 ENST00000380258.2 |
KIAA0101
|
KIAA0101 |
| chr10_-_90611566 | 0.06 |
ENST00000371930.4
|
ANKRD22
|
ankyrin repeat domain 22 |
| chr6_-_32977345 | 0.06 |
ENST00000450833.2
ENST00000374813.1 ENST00000229829.5 |
HLA-DOA
|
major histocompatibility complex, class II, DO alpha |
| chr3_-_47950745 | 0.05 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr10_-_95241951 | 0.05 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr3_-_55001115 | 0.05 |
ENST00000493075.1
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1 |
| chr5_-_169725231 | 0.05 |
ENST00000046794.5
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
| chr11_-_118047376 | 0.05 |
ENST00000278947.5
|
SCN2B
|
sodium channel, voltage-gated, type II, beta subunit |
| chr3_+_69812701 | 0.04 |
ENST00000472437.1
|
MITF
|
microphthalmia-associated transcription factor |
| chrM_+_8527 | 0.04 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase 6 |
| chr4_+_71337834 | 0.04 |
ENST00000304887.5
|
MUC7
|
mucin 7, secreted |
| chr8_-_37707356 | 0.04 |
ENST00000520601.1
ENST00000521170.1 ENST00000220659.6 |
BRF2
|
BRF2, RNA polymerase III transcription initiation factor 50 kDa subunit |
| chr20_-_32308028 | 0.04 |
ENST00000409299.3
ENST00000217398.3 ENST00000344022.3 |
PXMP4
|
peroxisomal membrane protein 4, 24kDa |
| chr12_-_772901 | 0.04 |
ENST00000305108.4
|
NINJ2
|
ninjurin 2 |
| chr17_+_62461569 | 0.03 |
ENST00000603557.1
ENST00000605096.1 |
MILR1
|
mast cell immunoglobulin-like receptor 1 |
| chrX_+_107288197 | 0.03 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr2_-_176046391 | 0.03 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr6_-_31125850 | 0.03 |
ENST00000507751.1
ENST00000448162.2 ENST00000502557.1 ENST00000503420.1 ENST00000507892.1 ENST00000507226.1 ENST00000513222.1 ENST00000503934.1 ENST00000396263.2 ENST00000508683.1 ENST00000428174.1 ENST00000448141.2 ENST00000507829.1 ENST00000455279.2 ENST00000376266.5 |
CCHCR1
|
coiled-coil alpha-helical rod protein 1 |
| chr1_-_115292591 | 0.03 |
ENST00000438362.2
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr8_+_41386761 | 0.03 |
ENST00000523277.2
|
GINS4
|
GINS complex subunit 4 (Sld5 homolog) |
| chr5_+_108083517 | 0.03 |
ENST00000281092.4
ENST00000536402.1 |
FER
|
fer (fps/fes related) tyrosine kinase |
| chr5_+_3596168 | 0.03 |
ENST00000302006.3
|
IRX1
|
iroquois homeobox 1 |
| chr5_+_179159813 | 0.03 |
ENST00000292599.3
|
MAML1
|
mastermind-like 1 (Drosophila) |
| chr12_-_10959892 | 0.03 |
ENST00000240615.2
|
TAS2R8
|
taste receptor, type 2, member 8 |
| chr3_+_113251143 | 0.02 |
ENST00000264852.4
ENST00000393830.3 |
SIDT1
|
SID1 transmembrane family, member 1 |
| chr19_-_14911023 | 0.02 |
ENST00000248073.2
|
OR7C1
|
olfactory receptor, family 7, subfamily C, member 1 |
| chr7_-_112635675 | 0.02 |
ENST00000447785.1
ENST00000451962.1 |
AC018464.3
|
AC018464.3 |
| chr1_-_120935894 | 0.02 |
ENST00000369383.4
ENST00000369384.4 |
FCGR1B
|
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
| chr14_+_39703084 | 0.02 |
ENST00000553728.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr6_+_26183958 | 0.02 |
ENST00000356530.3
|
HIST1H2BE
|
histone cluster 1, H2be |
| chr9_-_21142144 | 0.01 |
ENST00000380229.2
|
IFNW1
|
interferon, omega 1 |
| chr1_+_207038699 | 0.01 |
ENST00000367098.1
|
IL20
|
interleukin 20 |
| chr14_-_92198403 | 0.01 |
ENST00000553329.1
ENST00000256343.3 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr13_-_99630233 | 0.01 |
ENST00000376460.1
ENST00000442173.1 |
DOCK9
|
dedicator of cytokinesis 9 |
| chr11_+_61976137 | 0.01 |
ENST00000244930.4
|
SCGB2A1
|
secretoglobin, family 2A, member 1 |
| chr11_-_95657231 | 0.01 |
ENST00000409459.1
ENST00000352297.7 ENST00000393223.3 ENST00000346299.5 |
MTMR2
|
myotubularin related protein 2 |
| chrX_+_107288239 | 0.00 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr4_+_72204755 | 0.00 |
ENST00000512686.1
ENST00000340595.3 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr14_+_96949319 | 0.00 |
ENST00000554706.1
|
AK7
|
adenylate kinase 7 |
| chr6_-_89927151 | 0.00 |
ENST00000454853.2
|
GABRR1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr8_+_105352050 | 0.00 |
ENST00000297581.2
|
DCSTAMP
|
dendrocyte expressed seven transmembrane protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX11
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0014839 | myoblast migration involved in skeletal muscle regeneration(GO:0014839) |
| 0.3 | 1.3 | GO:1903282 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 2.7 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.9 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 0.5 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.1 | 0.8 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.8 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 1.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 0.4 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.2 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.1 | 0.3 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.5 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.2 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.2 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.3 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.5 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.2 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.5 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0071351 | interleukin-18-mediated signaling pathway(GO:0035655) cellular response to interleukin-18(GO:0071351) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.4 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.2 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 1.1 | GO:0046847 | filopodium assembly(GO:0046847) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.4 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 0.3 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 2.5 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 1.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.3 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.1 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.0 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.0 | 0.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.2 | 1.7 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.2 | 0.8 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.2 | 0.5 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.1 | 0.4 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.1 | 1.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.5 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.2 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 0.4 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.3 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.3 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.7 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.8 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.3 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 3.8 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |