Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
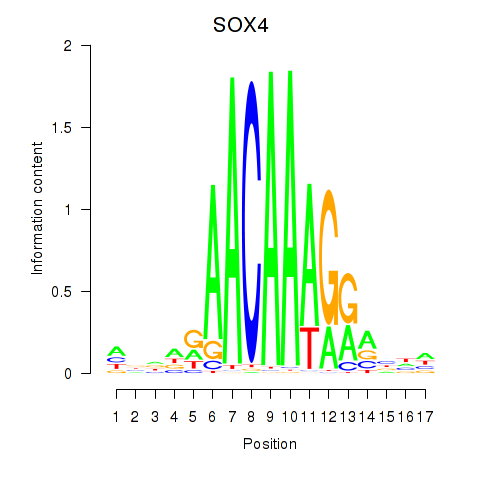
Results for SOX4
Z-value: 0.79
Transcription factors associated with SOX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX4
|
ENSG00000124766.4 | SRY-box transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX4 | hg19_v2_chr6_+_21593972_21594071 | 0.49 | 1.2e-02 | Click! |
Activity profile of SOX4 motif
Sorted Z-values of SOX4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_186877502 | 2.79 |
ENST00000431902.1
ENST00000284776.7 ENST00000415274.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr20_-_39317868 | 2.72 |
ENST00000373313.2
|
MAFB
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B |
| chr8_-_80993010 | 2.28 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr4_-_159080806 | 1.94 |
ENST00000590648.1
|
FAM198B
|
family with sequence similarity 198, member B |
| chr5_-_111091948 | 1.91 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr11_-_10829851 | 1.86 |
ENST00000532082.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr3_+_141105235 | 1.82 |
ENST00000503809.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr9_-_14313641 | 1.64 |
ENST00000380953.1
|
NFIB
|
nuclear factor I/B |
| chr12_+_93963590 | 1.52 |
ENST00000340600.2
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr16_+_30406721 | 1.49 |
ENST00000320159.2
|
ZNF48
|
zinc finger protein 48 |
| chr5_-_39425222 | 1.48 |
ENST00000320816.6
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chrX_-_63005405 | 1.44 |
ENST00000374878.1
ENST00000437457.2 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr5_-_39425068 | 1.39 |
ENST00000515700.1
ENST00000339788.6 |
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr5_-_39425290 | 1.36 |
ENST00000545653.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr2_+_33172221 | 1.35 |
ENST00000354476.3
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr11_-_10830463 | 1.30 |
ENST00000527419.1
ENST00000530211.1 ENST00000530702.1 ENST00000524932.1 ENST00000532570.1 |
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr1_+_97188188 | 1.29 |
ENST00000541987.1
|
PTBP2
|
polypyrimidine tract binding protein 2 |
| chr7_-_27219849 | 1.26 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr10_+_18948311 | 1.26 |
ENST00000377275.3
|
ARL5B
|
ADP-ribosylation factor-like 5B |
| chr8_-_17579726 | 1.25 |
ENST00000381861.3
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr9_-_14313893 | 1.25 |
ENST00000380921.3
ENST00000380959.3 |
NFIB
|
nuclear factor I/B |
| chr18_+_6729698 | 1.23 |
ENST00000383472.4
|
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr17_+_67498538 | 1.19 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr20_+_30598231 | 1.18 |
ENST00000300415.8
ENST00000262659.8 |
CCM2L
|
cerebral cavernous malformation 2-like |
| chr8_-_6420565 | 1.11 |
ENST00000338312.6
|
ANGPT2
|
angiopoietin 2 |
| chr1_-_85930246 | 1.09 |
ENST00000426972.3
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr15_+_25200108 | 1.07 |
ENST00000577949.1
ENST00000338094.6 ENST00000338327.4 ENST00000579070.1 ENST00000577565.1 |
SNURF
SNRPN
|
SNRPN upstream reading frame protein small nuclear ribonucleoprotein polypeptide N |
| chr7_+_116312411 | 1.03 |
ENST00000456159.1
ENST00000397752.3 ENST00000318493.6 |
MET
|
met proto-oncogene |
| chr17_+_25621102 | 1.01 |
ENST00000581440.1
ENST00000262394.2 ENST00000583742.1 ENST00000579733.1 ENST00000583193.1 ENST00000581185.1 ENST00000427287.2 ENST00000348811.2 |
WSB1
|
WD repeat and SOCS box containing 1 |
| chr17_-_14683517 | 1.01 |
ENST00000379640.1
|
AC005863.1
|
AC005863.1 |
| chr1_+_185703513 | 0.98 |
ENST00000271588.4
ENST00000367492.2 |
HMCN1
|
hemicentin 1 |
| chr15_+_25200074 | 0.96 |
ENST00000390687.4
ENST00000584968.1 ENST00000346403.6 ENST00000554227.2 |
SNRPN
|
small nuclear ribonucleoprotein polypeptide N |
| chr8_-_6420759 | 0.96 |
ENST00000523120.1
|
ANGPT2
|
angiopoietin 2 |
| chr13_+_39612442 | 0.90 |
ENST00000470258.1
ENST00000379600.3 |
NHLRC3
|
NHL repeat containing 3 |
| chr2_+_33172012 | 0.86 |
ENST00000404816.2
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr13_+_111855414 | 0.83 |
ENST00000375737.5
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr8_-_6420930 | 0.83 |
ENST00000325203.5
|
ANGPT2
|
angiopoietin 2 |
| chr8_-_6420777 | 0.82 |
ENST00000415216.1
|
ANGPT2
|
angiopoietin 2 |
| chr14_+_69658480 | 0.82 |
ENST00000409949.1
ENST00000409242.1 ENST00000312994.5 ENST00000413191.1 |
EXD2
|
exonuclease 3'-5' domain containing 2 |
| chr4_+_78079450 | 0.79 |
ENST00000395640.1
ENST00000512918.1 |
CCNG2
|
cyclin G2 |
| chr4_+_78079570 | 0.77 |
ENST00000509972.1
|
CCNG2
|
cyclin G2 |
| chr14_+_91580777 | 0.74 |
ENST00000525393.2
ENST00000428926.2 ENST00000517362.1 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr14_+_91581011 | 0.74 |
ENST00000523894.1
ENST00000522322.1 ENST00000523771.1 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr1_+_66458072 | 0.73 |
ENST00000423207.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr2_-_106015527 | 0.73 |
ENST00000344213.4
ENST00000358129.4 |
FHL2
|
four and a half LIM domains 2 |
| chr14_+_75746781 | 0.72 |
ENST00000555347.1
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr14_+_91580708 | 0.67 |
ENST00000518868.1
|
C14orf159
|
chromosome 14 open reading frame 159 |
| chr9_-_74979420 | 0.64 |
ENST00000343431.2
ENST00000376956.3 |
ZFAND5
|
zinc finger, AN1-type domain 5 |
| chr7_+_115850547 | 0.63 |
ENST00000358204.4
ENST00000455989.1 ENST00000537767.1 |
TES
|
testis derived transcript (3 LIM domains) |
| chr9_-_16727978 | 0.61 |
ENST00000418777.1
ENST00000468187.2 |
BNC2
|
basonuclin 2 |
| chr5_+_140739537 | 0.59 |
ENST00000522605.1
|
PCDHGB2
|
protocadherin gamma subfamily B, 2 |
| chr7_-_19813192 | 0.59 |
ENST00000422233.1
ENST00000433641.1 |
TMEM196
|
transmembrane protein 196 |
| chr11_+_46402744 | 0.57 |
ENST00000533952.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr10_-_18948156 | 0.57 |
ENST00000414939.1
ENST00000449529.1 ENST00000456217.1 ENST00000444660.1 |
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr18_+_6729725 | 0.56 |
ENST00000400091.2
ENST00000583410.1 ENST00000584387.1 |
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr22_+_27017921 | 0.56 |
ENST00000354760.3
|
CRYBA4
|
crystallin, beta A4 |
| chr5_+_140579162 | 0.55 |
ENST00000536699.1
ENST00000354757.3 |
PCDHB11
|
protocadherin beta 11 |
| chrX_-_2847366 | 0.55 |
ENST00000381154.1
|
ARSD
|
arylsulfatase D |
| chr1_+_207002222 | 0.54 |
ENST00000270218.6
|
IL19
|
interleukin 19 |
| chr4_+_78078304 | 0.52 |
ENST00000316355.5
ENST00000354403.5 ENST00000502280.1 |
CCNG2
|
cyclin G2 |
| chr19_+_1249869 | 0.52 |
ENST00000591446.2
|
MIDN
|
midnolin |
| chr1_+_81771806 | 0.50 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr2_+_208423840 | 0.50 |
ENST00000539789.1
|
CREB1
|
cAMP responsive element binding protein 1 |
| chr14_+_24521202 | 0.49 |
ENST00000334420.7
ENST00000342740.5 |
LRRC16B
|
leucine rich repeat containing 16B |
| chr1_+_228337553 | 0.49 |
ENST00000366714.2
|
GJC2
|
gap junction protein, gamma 2, 47kDa |
| chr12_+_132379160 | 0.49 |
ENST00000321867.4
|
ULK1
|
unc-51 like autophagy activating kinase 1 |
| chr19_-_17375527 | 0.49 |
ENST00000431146.2
ENST00000594190.1 |
USHBP1
|
Usher syndrome 1C binding protein 1 |
| chr19_+_10765699 | 0.48 |
ENST00000590009.1
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chr9_+_91003271 | 0.47 |
ENST00000375859.3
ENST00000541629.1 |
SPIN1
|
spindlin 1 |
| chr14_-_51027838 | 0.45 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr15_+_65823092 | 0.44 |
ENST00000566074.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr5_+_140749803 | 0.43 |
ENST00000576222.1
|
PCDHGB3
|
protocadherin gamma subfamily B, 3 |
| chr15_+_65822756 | 0.43 |
ENST00000562901.1
ENST00000261875.5 ENST00000442729.2 ENST00000565299.1 ENST00000568793.1 |
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr7_-_148581360 | 0.43 |
ENST00000320356.2
ENST00000541220.1 ENST00000483967.1 ENST00000536783.1 |
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr7_-_107643674 | 0.42 |
ENST00000222399.6
|
LAMB1
|
laminin, beta 1 |
| chr12_+_32260085 | 0.42 |
ENST00000548411.1
ENST00000281474.5 ENST00000551086.1 |
BICD1
|
bicaudal D homolog 1 (Drosophila) |
| chr18_+_56530794 | 0.42 |
ENST00000590285.1
ENST00000586085.1 ENST00000589288.1 |
ZNF532
|
zinc finger protein 532 |
| chr16_+_14980632 | 0.41 |
ENST00000565655.1
|
NOMO1
|
NODAL modulator 1 |
| chr5_+_102201509 | 0.40 |
ENST00000348126.2
ENST00000379787.4 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr20_+_45338126 | 0.40 |
ENST00000359271.2
|
SLC2A10
|
solute carrier family 2 (facilitated glucose transporter), member 10 |
| chr7_-_148581251 | 0.39 |
ENST00000478654.1
ENST00000460911.1 ENST00000350995.2 |
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr11_-_62414070 | 0.39 |
ENST00000540933.1
ENST00000346178.4 ENST00000356638.3 ENST00000534779.1 ENST00000525994.1 |
GANAB
|
glucosidase, alpha; neutral AB |
| chr20_-_17511962 | 0.38 |
ENST00000377873.3
|
BFSP1
|
beaded filament structural protein 1, filensin |
| chr2_+_208423891 | 0.37 |
ENST00000448277.1
ENST00000457101.1 |
CREB1
|
cAMP responsive element binding protein 1 |
| chr8_-_95908902 | 0.36 |
ENST00000520509.1
|
CCNE2
|
cyclin E2 |
| chr11_-_94965667 | 0.35 |
ENST00000542176.1
ENST00000278499.2 |
SESN3
|
sestrin 3 |
| chr15_-_34502197 | 0.34 |
ENST00000557877.1
|
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr18_+_7946941 | 0.34 |
ENST00000444013.1
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr22_-_31688381 | 0.32 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr4_-_22444733 | 0.31 |
ENST00000508133.1
|
GPR125
|
G protein-coupled receptor 125 |
| chr4_+_106629929 | 0.31 |
ENST00000512828.1
ENST00000394730.3 ENST00000507281.1 ENST00000515279.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
| chr10_+_22605374 | 0.31 |
ENST00000448361.1
|
COMMD3
|
COMM domain containing 3 |
| chr7_+_134551583 | 0.31 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr10_+_22605304 | 0.31 |
ENST00000475460.2
ENST00000602390.1 ENST00000489125.2 ENST00000456711.1 ENST00000444869.1 |
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 readthrough COMM domain containing 3 |
| chr14_+_69658194 | 0.31 |
ENST00000409018.3
ENST00000409014.1 ENST00000409675.1 |
EXD2
|
exonuclease 3'-5' domain containing 2 |
| chr19_+_1248547 | 0.30 |
ENST00000586757.1
ENST00000300952.2 |
MIDN
|
midnolin |
| chr1_+_150039369 | 0.30 |
ENST00000369130.3
ENST00000369128.5 |
VPS45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr5_+_102201430 | 0.29 |
ENST00000438793.3
ENST00000346918.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chrX_-_24690771 | 0.29 |
ENST00000379145.1
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr7_+_139528952 | 0.28 |
ENST00000416849.2
ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr14_+_101299520 | 0.28 |
ENST00000455531.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr19_-_17375541 | 0.28 |
ENST00000252597.3
|
USHBP1
|
Usher syndrome 1C binding protein 1 |
| chr1_-_93645818 | 0.27 |
ENST00000370280.1
ENST00000479918.1 |
TMED5
|
transmembrane emp24 protein transport domain containing 5 |
| chr11_-_6677018 | 0.27 |
ENST00000299441.3
|
DCHS1
|
dachsous cadherin-related 1 |
| chr14_-_31676674 | 0.26 |
ENST00000399332.1
ENST00000556224.1 |
HECTD1
|
HECT domain containing E3 ubiquitin protein ligase 1 |
| chr3_+_50649302 | 0.26 |
ENST00000446044.1
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr14_-_20923195 | 0.26 |
ENST00000206542.4
|
OSGEP
|
O-sialoglycoprotein endopeptidase |
| chr8_-_16043780 | 0.25 |
ENST00000445506.2
|
MSR1
|
macrophage scavenger receptor 1 |
| chr6_-_132834184 | 0.25 |
ENST00000367941.2
ENST00000367937.4 |
STX7
|
syntaxin 7 |
| chr14_-_73493784 | 0.24 |
ENST00000553891.1
|
ZFYVE1
|
zinc finger, FYVE domain containing 1 |
| chr14_+_50234827 | 0.23 |
ENST00000554589.1
ENST00000557247.1 |
KLHDC2
|
kelch domain containing 2 |
| chr5_+_93954039 | 0.22 |
ENST00000265140.5
|
ANKRD32
|
ankyrin repeat domain 32 |
| chr1_-_32801825 | 0.22 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr4_-_69111401 | 0.21 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr4_+_71588372 | 0.21 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr1_-_26233423 | 0.21 |
ENST00000357865.2
|
STMN1
|
stathmin 1 |
| chr14_+_91580732 | 0.21 |
ENST00000519019.1
ENST00000523816.1 ENST00000517518.1 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr3_+_159570722 | 0.20 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr1_+_93645314 | 0.20 |
ENST00000343253.7
|
CCDC18
|
coiled-coil domain containing 18 |
| chr12_+_109915179 | 0.20 |
ENST00000434735.2
|
UBE3B
|
ubiquitin protein ligase E3B |
| chr16_-_31085033 | 0.19 |
ENST00000414399.1
|
ZNF668
|
zinc finger protein 668 |
| chrX_-_73512411 | 0.19 |
ENST00000602576.1
ENST00000429124.1 |
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr17_+_59489112 | 0.18 |
ENST00000335108.2
|
C17orf82
|
chromosome 17 open reading frame 82 |
| chr4_+_2819883 | 0.18 |
ENST00000511747.1
ENST00000503393.2 |
SH3BP2
|
SH3-domain binding protein 2 |
| chrX_+_51927919 | 0.18 |
ENST00000416960.1
|
MAGED4
|
melanoma antigen family D, 4 |
| chr16_+_72088376 | 0.18 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr3_-_50649192 | 0.18 |
ENST00000443053.2
ENST00000348721.3 |
CISH
|
cytokine inducible SH2-containing protein |
| chr14_-_73493825 | 0.18 |
ENST00000318876.5
ENST00000556143.1 |
ZFYVE1
|
zinc finger, FYVE domain containing 1 |
| chrX_-_135962876 | 0.17 |
ENST00000431446.3
ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX
|
RNA binding motif protein, X-linked |
| chr12_-_10282742 | 0.17 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr15_-_45670924 | 0.17 |
ENST00000396659.3
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr1_+_28261492 | 0.16 |
ENST00000373894.3
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr12_-_48963829 | 0.16 |
ENST00000301046.2
ENST00000549817.1 |
LALBA
|
lactalbumin, alpha- |
| chr7_-_35293740 | 0.16 |
ENST00000408931.3
|
TBX20
|
T-box 20 |
| chr21_+_30671690 | 0.15 |
ENST00000399921.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr9_-_39239171 | 0.15 |
ENST00000358144.2
|
CNTNAP3
|
contactin associated protein-like 3 |
| chrX_+_36053908 | 0.15 |
ENST00000378660.2
|
CHDC2
|
calponin homology domain containing 2 |
| chr1_+_28844778 | 0.15 |
ENST00000411533.1
|
RCC1
|
regulator of chromosome condensation 1 |
| chr2_+_223536428 | 0.15 |
ENST00000446656.3
|
MOGAT1
|
monoacylglycerol O-acyltransferase 1 |
| chr5_+_140480083 | 0.14 |
ENST00000231130.2
|
PCDHB3
|
protocadherin beta 3 |
| chr1_+_109419596 | 0.14 |
ENST00000435987.1
ENST00000264126.3 |
GPSM2
|
G-protein signaling modulator 2 |
| chr9_+_124088860 | 0.14 |
ENST00000373806.1
|
GSN
|
gelsolin |
| chr3_+_57875711 | 0.14 |
ENST00000442599.2
|
SLMAP
|
sarcolemma associated protein |
| chr15_+_75940218 | 0.14 |
ENST00000308527.5
|
SNX33
|
sorting nexin 33 |
| chr8_+_9911778 | 0.13 |
ENST00000317173.4
ENST00000441698.2 |
MSRA
|
methionine sulfoxide reductase A |
| chr4_+_159690218 | 0.13 |
ENST00000264433.6
|
FNIP2
|
folliculin interacting protein 2 |
| chr7_-_42276612 | 0.13 |
ENST00000395925.3
ENST00000437480.1 |
GLI3
|
GLI family zinc finger 3 |
| chr12_-_10282836 | 0.13 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr1_+_28261621 | 0.13 |
ENST00000549094.1
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr1_+_209602771 | 0.13 |
ENST00000440276.1
|
MIR205HG
|
MIR205 host gene (non-protein coding) |
| chr3_-_197024965 | 0.13 |
ENST00000392382.2
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr22_-_31688431 | 0.13 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chrX_+_70503433 | 0.13 |
ENST00000276079.8
ENST00000373856.3 ENST00000373841.1 ENST00000420903.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr20_+_18125727 | 0.12 |
ENST00000489634.2
|
CSRP2BP
|
CSRP2 binding protein |
| chr20_+_58533471 | 0.12 |
ENST00000244047.5
ENST00000348616.4 |
CDH26
|
cadherin 26 |
| chr12_-_122296755 | 0.12 |
ENST00000289004.4
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase |
| chr5_+_140767452 | 0.12 |
ENST00000519479.1
|
PCDHGB4
|
protocadherin gamma subfamily B, 4 |
| chr12_-_95945246 | 0.12 |
ENST00000258499.3
|
USP44
|
ubiquitin specific peptidase 44 |
| chr1_+_153940713 | 0.12 |
ENST00000368601.1
ENST00000368603.1 ENST00000368600.3 |
CREB3L4
|
cAMP responsive element binding protein 3-like 4 |
| chr3_-_99833333 | 0.12 |
ENST00000354552.3
ENST00000331335.5 ENST00000398326.2 |
FILIP1L
|
filamin A interacting protein 1-like |
| chr12_+_8666126 | 0.12 |
ENST00000299665.2
|
CLEC4D
|
C-type lectin domain family 4, member D |
| chr4_-_54424366 | 0.11 |
ENST00000306888.2
|
LNX1
|
ligand of numb-protein X 1, E3 ubiquitin protein ligase |
| chr3_-_197024394 | 0.11 |
ENST00000434148.1
ENST00000412364.2 ENST00000436682.1 ENST00000456699.2 ENST00000392380.2 |
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr2_-_20425158 | 0.11 |
ENST00000381150.1
|
SDC1
|
syndecan 1 |
| chr17_-_10372875 | 0.11 |
ENST00000255381.2
|
MYH4
|
myosin, heavy chain 4, skeletal muscle |
| chrX_+_12993202 | 0.11 |
ENST00000451311.2
ENST00000380636.1 |
TMSB4X
|
thymosin beta 4, X-linked |
| chr1_+_160147176 | 0.10 |
ENST00000470705.1
|
ATP1A4
|
ATPase, Na+/K+ transporting, alpha 4 polypeptide |
| chr17_-_72358001 | 0.10 |
ENST00000375366.3
|
BTBD17
|
BTB (POZ) domain containing 17 |
| chr1_+_153940346 | 0.10 |
ENST00000405694.3
ENST00000449724.1 ENST00000368607.3 ENST00000271889.4 |
CREB3L4
|
cAMP responsive element binding protein 3-like 4 |
| chr16_+_524850 | 0.10 |
ENST00000450428.1
ENST00000452814.1 |
RAB11FIP3
|
RAB11 family interacting protein 3 (class II) |
| chr1_-_152297679 | 0.10 |
ENST00000368799.1
|
FLG
|
filaggrin |
| chr1_-_115259337 | 0.10 |
ENST00000369535.4
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr17_+_57297807 | 0.10 |
ENST00000284116.4
ENST00000581140.1 ENST00000581276.1 |
GDPD1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr12_-_10282681 | 0.10 |
ENST00000533022.1
|
CLEC7A
|
C-type lectin domain family 7, member A |
| chr7_-_150946015 | 0.10 |
ENST00000262188.8
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr19_+_49128209 | 0.10 |
ENST00000599748.1
ENST00000443164.1 ENST00000599029.1 |
SPHK2
|
sphingosine kinase 2 |
| chr11_-_61560053 | 0.09 |
ENST00000537328.1
|
TMEM258
|
transmembrane protein 258 |
| chrX_-_15683147 | 0.09 |
ENST00000380342.3
|
TMEM27
|
transmembrane protein 27 |
| chr5_+_140787600 | 0.09 |
ENST00000520790.1
|
PCDHGB6
|
protocadherin gamma subfamily B, 6 |
| chr14_+_20923350 | 0.09 |
ENST00000555414.1
ENST00000216714.3 ENST00000553681.1 ENST00000557344.1 ENST00000398030.4 ENST00000557181.1 ENST00000555839.1 ENST00000553368.1 ENST00000556054.1 ENST00000557054.1 ENST00000557592.1 ENST00000557150.1 |
APEX1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr9_+_139221880 | 0.09 |
ENST00000392945.3
ENST00000440944.1 |
GPSM1
|
G-protein signaling modulator 1 |
| chr17_+_9066252 | 0.08 |
ENST00000436734.1
|
NTN1
|
netrin 1 |
| chr6_-_27440460 | 0.07 |
ENST00000377419.1
|
ZNF184
|
zinc finger protein 184 |
| chr6_+_43265992 | 0.07 |
ENST00000449231.1
ENST00000372589.3 ENST00000372585.5 |
SLC22A7
|
solute carrier family 22 (organic anion transporter), member 7 |
| chr1_+_28261533 | 0.07 |
ENST00000411604.1
ENST00000373888.4 |
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr13_-_41593425 | 0.07 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr1_-_155880672 | 0.07 |
ENST00000609492.1
ENST00000368322.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr12_+_55248289 | 0.07 |
ENST00000308796.6
|
MUCL1
|
mucin-like 1 |
| chr7_+_30185406 | 0.07 |
ENST00000324489.5
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr14_+_24439148 | 0.07 |
ENST00000543805.1
ENST00000534993.1 |
DHRS4L2
|
dehydrogenase/reductase (SDR family) member 4 like 2 |
| chr15_+_81293254 | 0.07 |
ENST00000267984.2
|
MESDC1
|
mesoderm development candidate 1 |
| chr17_+_64961026 | 0.06 |
ENST00000262138.3
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4 |
| chr2_-_127977654 | 0.06 |
ENST00000409327.1
|
CYP27C1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr6_+_123038689 | 0.06 |
ENST00000354275.2
ENST00000368446.1 |
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr1_-_1243252 | 0.06 |
ENST00000353662.3
|
ACAP3
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
| chr1_-_182642017 | 0.05 |
ENST00000367557.4
ENST00000258302.4 |
RGS8
|
regulator of G-protein signaling 8 |
| chr16_+_6069072 | 0.05 |
ENST00000547605.1
ENST00000550418.1 ENST00000553186.1 |
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr1_-_167883327 | 0.05 |
ENST00000476818.2
ENST00000367851.4 ENST00000367848.1 |
ADCY10
|
adenylate cyclase 10 (soluble) |
| chr10_+_51187938 | 0.05 |
ENST00000311663.5
|
FAM21D
|
family with sequence similarity 21, member D |
| chrX_+_12993336 | 0.05 |
ENST00000380635.1
|
TMSB4X
|
thymosin beta 4, X-linked |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.2 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.9 | 3.7 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.9 | 2.7 | GO:0035284 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.6 | 1.8 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.4 | 2.9 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.4 | 1.2 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.4 | 1.2 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.3 | 0.8 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.2 | 1.2 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.2 | 2.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.2 | 0.7 | GO:0018032 | protein amidation(GO:0018032) |
| 0.1 | 0.7 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.9 | GO:0034670 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.1 | 0.4 | GO:1900276 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.1 | 0.8 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.4 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.1 | 0.6 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 0.8 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 0.3 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.1 | 0.7 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.4 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.2 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 1.1 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.1 | 1.5 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 2.3 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.7 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.5 | GO:0075071 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.1 | 0.2 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.1 | 0.2 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.1 | 0.2 | GO:0003193 | pulmonary valve formation(GO:0003193) visceral motor neuron differentiation(GO:0021524) foramen ovale closure(GO:0035922) |
| 0.1 | 0.2 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 2.4 | GO:0006536 | glutamate metabolic process(GO:0006536) |
| 0.0 | 0.1 | GO:0048627 | myoblast development(GO:0048627) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 1.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 1.5 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 3.2 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 1.4 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.1 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.4 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.4 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.2 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.5 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 | 0.1 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.4 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.3 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.6 | GO:0003416 | endochondral bone growth(GO:0003416) bone growth(GO:0098868) |
| 0.0 | 0.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.6 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 0.5 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.2 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.4 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 0.9 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.2 | 2.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 3.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.4 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.1 | 0.4 | GO:0039713 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.1 | 0.9 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 2.0 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 0.4 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 0.7 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 2.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.8 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.8 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 1.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 3.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 1.1 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.4 | 2.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.3 | 1.5 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.2 | 4.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.2 | 0.7 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.2 | 1.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.4 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 1.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 3.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.9 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.1 | 0.3 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.1 | 0.8 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.5 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 1.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.4 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 3.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 3.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.5 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.7 | GO:0043015 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.1 | GO:0005503 | all-trans retinal binding(GO:0005503) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.4 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 1.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.3 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 2.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.6 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.9 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 4.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 3.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 3.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 2.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.8 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.0 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 3.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.9 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 1.4 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 1.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.7 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.9 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 3.2 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 2.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.0 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |