Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
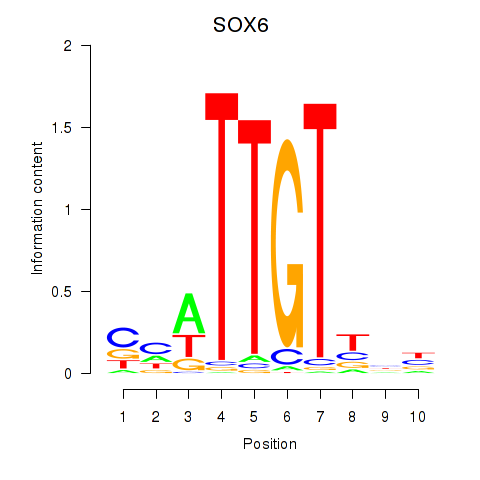
Results for SOX6
Z-value: 0.50
Transcription factors associated with SOX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX6
|
ENSG00000110693.11 | SRY-box transcription factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX6 | hg19_v2_chr11_-_16430399_16430440 | 0.13 | 5.3e-01 | Click! |
Activity profile of SOX6 motif
Sorted Z-values of SOX6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_208031542 | 1.24 |
ENST00000423015.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr4_+_41614720 | 1.19 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr4_+_41614909 | 1.16 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr14_-_51027838 | 1.12 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr11_-_10830463 | 0.97 |
ENST00000527419.1
ENST00000530211.1 ENST00000530702.1 ENST00000524932.1 ENST00000532570.1 |
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr14_+_103589789 | 0.80 |
ENST00000558056.1
ENST00000560869.1 |
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr6_+_126240442 | 0.62 |
ENST00000448104.1
ENST00000438495.1 ENST00000444128.1 |
NCOA7
|
nuclear receptor coactivator 7 |
| chr20_+_6748311 | 0.61 |
ENST00000378827.4
|
BMP2
|
bone morphogenetic protein 2 |
| chr2_-_37899323 | 0.55 |
ENST00000295324.3
ENST00000457889.1 |
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr1_+_182808474 | 0.55 |
ENST00000367549.3
|
DHX9
|
DEAH (Asp-Glu-Ala-His) box helicase 9 |
| chrX_-_117107542 | 0.50 |
ENST00000371878.1
|
KLHL13
|
kelch-like family member 13 |
| chr18_+_6729698 | 0.50 |
ENST00000383472.4
|
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr10_+_35416223 | 0.50 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr14_+_22748980 | 0.44 |
ENST00000390465.2
|
TRAV38-2DV8
|
T cell receptor alpha variable 38-2/delta variable 8 |
| chr22_-_27014043 | 0.43 |
ENST00000215939.2
|
CRYBB1
|
crystallin, beta B1 |
| chr8_-_6420777 | 0.43 |
ENST00000415216.1
|
ANGPT2
|
angiopoietin 2 |
| chr4_+_144303093 | 0.41 |
ENST00000505913.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr7_+_77167343 | 0.38 |
ENST00000433369.2
ENST00000415482.2 |
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr11_+_128563948 | 0.38 |
ENST00000534087.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr2_+_111880242 | 0.37 |
ENST00000393252.3
|
BCL2L11
|
BCL2-like 11 (apoptosis facilitator) |
| chr21_+_33784670 | 0.37 |
ENST00000300255.2
|
EVA1C
|
eva-1 homolog C (C. elegans) |
| chr14_-_35344093 | 0.36 |
ENST00000382422.2
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A |
| chr11_+_128563652 | 0.35 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr6_+_132455118 | 0.33 |
ENST00000458028.1
|
LINC01013
|
long intergenic non-protein coding RNA 1013 |
| chr17_-_18585131 | 0.33 |
ENST00000443457.1
ENST00000583002.1 |
ZNF286B
|
zinc finger protein 286B |
| chr9_-_74979420 | 0.33 |
ENST00000343431.2
ENST00000376956.3 |
ZFAND5
|
zinc finger, AN1-type domain 5 |
| chr9_-_123476719 | 0.33 |
ENST00000373930.3
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr1_+_228337553 | 0.32 |
ENST00000366714.2
|
GJC2
|
gap junction protein, gamma 2, 47kDa |
| chr17_-_76719807 | 0.30 |
ENST00000589297.1
|
CYTH1
|
cytohesin 1 |
| chr6_-_75915757 | 0.30 |
ENST00000322507.8
|
COL12A1
|
collagen, type XII, alpha 1 |
| chr5_+_140749803 | 0.30 |
ENST00000576222.1
|
PCDHGB3
|
protocadherin gamma subfamily B, 3 |
| chrX_+_84499081 | 0.29 |
ENST00000276123.3
|
ZNF711
|
zinc finger protein 711 |
| chr17_-_76713100 | 0.28 |
ENST00000585509.1
|
CYTH1
|
cytohesin 1 |
| chr16_+_30406721 | 0.28 |
ENST00000320159.2
|
ZNF48
|
zinc finger protein 48 |
| chr15_-_82338460 | 0.28 |
ENST00000558133.1
ENST00000329713.4 |
MEX3B
|
mex-3 RNA binding family member B |
| chrX_-_45060135 | 0.27 |
ENST00000398000.2
ENST00000377934.4 |
CXorf36
|
chromosome X open reading frame 36 |
| chr15_-_73075964 | 0.27 |
ENST00000563907.1
|
ADPGK
|
ADP-dependent glucokinase |
| chr4_+_55524085 | 0.26 |
ENST00000412167.2
ENST00000288135.5 |
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
| chr10_-_25241499 | 0.26 |
ENST00000376378.1
ENST00000376376.3 ENST00000320152.6 |
PRTFDC1
|
phosphoribosyl transferase domain containing 1 |
| chr15_+_81293254 | 0.25 |
ENST00000267984.2
|
MESDC1
|
mesoderm development candidate 1 |
| chr12_-_109219937 | 0.25 |
ENST00000546697.1
|
SSH1
|
slingshot protein phosphatase 1 |
| chr15_+_63335899 | 0.24 |
ENST00000561266.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr4_+_54243917 | 0.24 |
ENST00000507166.1
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr15_+_85923797 | 0.24 |
ENST00000559362.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr15_+_52311398 | 0.23 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr17_-_40288449 | 0.23 |
ENST00000552162.1
ENST00000550504.1 |
RAB5C
|
RAB5C, member RAS oncogene family |
| chr2_-_231989808 | 0.23 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr12_+_69979446 | 0.23 |
ENST00000543146.2
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr2_+_10262857 | 0.23 |
ENST00000304567.5
|
RRM2
|
ribonucleotide reductase M2 |
| chr14_+_23791159 | 0.22 |
ENST00000557702.1
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr6_+_132455526 | 0.22 |
ENST00000443303.1
|
LINC01013
|
long intergenic non-protein coding RNA 1013 |
| chr3_-_65583561 | 0.22 |
ENST00000460329.2
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr13_+_24144796 | 0.21 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr7_-_128415844 | 0.21 |
ENST00000249389.2
|
OPN1SW
|
opsin 1 (cone pigments), short-wave-sensitive |
| chr1_-_182360498 | 0.21 |
ENST00000417584.2
|
GLUL
|
glutamate-ammonia ligase |
| chr19_+_47105309 | 0.21 |
ENST00000599839.1
ENST00000596362.1 |
CALM3
|
calmodulin 3 (phosphorylase kinase, delta) |
| chrX_+_70503433 | 0.21 |
ENST00000276079.8
ENST00000373856.3 ENST00000373841.1 ENST00000420903.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr11_-_69519410 | 0.21 |
ENST00000294312.3
|
FGF19
|
fibroblast growth factor 19 |
| chr9_+_139874683 | 0.21 |
ENST00000444903.1
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain) |
| chr16_+_23765948 | 0.21 |
ENST00000300113.2
|
CHP2
|
calcineurin-like EF-hand protein 2 |
| chr5_-_180018540 | 0.20 |
ENST00000292641.3
|
SCGB3A1
|
secretoglobin, family 3A, member 1 |
| chr4_-_146101304 | 0.20 |
ENST00000447906.2
|
OTUD4
|
OTU domain containing 4 |
| chr12_+_93096619 | 0.20 |
ENST00000397833.3
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr1_+_24286287 | 0.20 |
ENST00000334351.7
ENST00000374468.1 |
PNRC2
|
proline-rich nuclear receptor coactivator 2 |
| chr5_-_146435694 | 0.20 |
ENST00000356826.3
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr2_-_158182410 | 0.19 |
ENST00000419116.2
ENST00000410096.1 |
ERMN
|
ermin, ERM-like protein |
| chr12_+_93096759 | 0.19 |
ENST00000544406.2
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr10_+_114043493 | 0.19 |
ENST00000369422.3
|
TECTB
|
tectorin beta |
| chr18_-_52989217 | 0.19 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr12_-_88974236 | 0.19 |
ENST00000228280.5
ENST00000552044.1 ENST00000357116.4 |
KITLG
|
KIT ligand |
| chr15_+_74610894 | 0.18 |
ENST00000558821.1
ENST00000268082.4 |
CCDC33
|
coiled-coil domain containing 33 |
| chr8_+_107738343 | 0.18 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr3_-_197024394 | 0.18 |
ENST00000434148.1
ENST00000412364.2 ENST00000436682.1 ENST00000456699.2 ENST00000392380.2 |
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr5_-_146435572 | 0.18 |
ENST00000394414.1
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr5_-_146435501 | 0.18 |
ENST00000336640.6
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr12_-_71003568 | 0.18 |
ENST00000547715.1
ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr14_-_71107921 | 0.17 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr1_-_201476274 | 0.17 |
ENST00000340006.2
|
CSRP1
|
cysteine and glycine-rich protein 1 |
| chr2_+_170590321 | 0.17 |
ENST00000392647.2
|
KLHL23
|
kelch-like family member 23 |
| chr4_+_71570430 | 0.17 |
ENST00000417478.2
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr11_+_34654011 | 0.17 |
ENST00000531794.1
|
EHF
|
ets homologous factor |
| chrX_+_135579238 | 0.17 |
ENST00000535601.1
ENST00000448450.1 ENST00000425695.1 |
HTATSF1
|
HIV-1 Tat specific factor 1 |
| chrX_+_84498989 | 0.17 |
ENST00000395402.1
|
ZNF711
|
zinc finger protein 711 |
| chr10_+_35415851 | 0.16 |
ENST00000374726.3
|
CREM
|
cAMP responsive element modulator |
| chr5_-_16936340 | 0.16 |
ENST00000507288.1
ENST00000513610.1 |
MYO10
|
myosin X |
| chr7_-_19813192 | 0.16 |
ENST00000422233.1
ENST00000433641.1 |
TMEM196
|
transmembrane protein 196 |
| chr4_+_26585538 | 0.16 |
ENST00000264866.4
|
TBC1D19
|
TBC1 domain family, member 19 |
| chr19_+_5681011 | 0.16 |
ENST00000581893.1
ENST00000411793.2 ENST00000301382.4 ENST00000581773.1 ENST00000423665.2 ENST00000583928.1 ENST00000342970.2 ENST00000422535.2 ENST00000581521.1 ENST00000339423.2 |
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like |
| chr16_-_31085033 | 0.16 |
ENST00000414399.1
|
ZNF668
|
zinc finger protein 668 |
| chr8_-_141810634 | 0.16 |
ENST00000521986.1
ENST00000523539.1 ENST00000538769.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr6_-_107436473 | 0.16 |
ENST00000369042.1
|
BEND3
|
BEN domain containing 3 |
| chr9_-_39239171 | 0.16 |
ENST00000358144.2
|
CNTNAP3
|
contactin associated protein-like 3 |
| chr18_+_6729725 | 0.16 |
ENST00000400091.2
ENST00000583410.1 ENST00000584387.1 |
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr11_-_76155618 | 0.16 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr7_+_116312411 | 0.16 |
ENST00000456159.1
ENST00000397752.3 ENST00000318493.6 |
MET
|
met proto-oncogene |
| chr17_-_14683517 | 0.15 |
ENST00000379640.1
|
AC005863.1
|
AC005863.1 |
| chrX_+_84499038 | 0.15 |
ENST00000373165.3
|
ZNF711
|
zinc finger protein 711 |
| chr7_-_99699538 | 0.15 |
ENST00000343023.6
ENST00000303887.5 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr17_+_36584662 | 0.15 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr12_+_100041527 | 0.15 |
ENST00000324341.1
|
FAM71C
|
family with sequence similarity 71, member C |
| chr12_+_104359576 | 0.15 |
ENST00000392872.3
ENST00000436021.2 |
TDG
|
thymine-DNA glycosylase |
| chr6_-_89927151 | 0.14 |
ENST00000454853.2
|
GABRR1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr11_-_82708435 | 0.14 |
ENST00000525117.1
ENST00000532548.1 |
RAB30
|
RAB30, member RAS oncogene family |
| chr9_+_124103625 | 0.14 |
ENST00000594963.1
|
AL161784.1
|
Uncharacterized protein |
| chr7_+_29234101 | 0.14 |
ENST00000435288.2
|
CHN2
|
chimerin 2 |
| chr7_+_29234028 | 0.14 |
ENST00000222792.6
|
CHN2
|
chimerin 2 |
| chrX_+_80457442 | 0.14 |
ENST00000373212.5
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like |
| chr11_-_82708519 | 0.14 |
ENST00000534301.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr16_+_2083265 | 0.14 |
ENST00000565855.1
ENST00000566198.1 |
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr1_-_201123546 | 0.13 |
ENST00000435310.1
ENST00000485839.2 ENST00000367330.1 |
TMEM9
|
transmembrane protein 9 |
| chr5_+_140593509 | 0.13 |
ENST00000341948.4
|
PCDHB13
|
protocadherin beta 13 |
| chr12_+_60058458 | 0.13 |
ENST00000548610.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr7_+_134464376 | 0.13 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr7_+_23146271 | 0.13 |
ENST00000545771.1
|
KLHL7
|
kelch-like family member 7 |
| chr12_-_109915098 | 0.13 |
ENST00000542858.1
ENST00000542262.1 ENST00000424763.2 |
KCTD10
|
potassium channel tetramerization domain containing 10 |
| chr4_+_78078304 | 0.13 |
ENST00000316355.5
ENST00000354403.5 ENST00000502280.1 |
CCNG2
|
cyclin G2 |
| chr16_+_3704822 | 0.13 |
ENST00000414110.2
|
DNASE1
|
deoxyribonuclease I |
| chr1_-_201123586 | 0.13 |
ENST00000414605.2
ENST00000367334.5 ENST00000367332.1 |
TMEM9
|
transmembrane protein 9 |
| chr4_+_54243798 | 0.13 |
ENST00000337488.6
ENST00000358575.5 ENST00000507922.1 |
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr4_+_26585686 | 0.13 |
ENST00000505206.1
ENST00000511789.1 |
TBC1D19
|
TBC1 domain family, member 19 |
| chr1_+_147374915 | 0.12 |
ENST00000240986.4
|
GJA8
|
gap junction protein, alpha 8, 50kDa |
| chr10_+_115511213 | 0.12 |
ENST00000361048.1
|
PLEKHS1
|
pleckstrin homology domain containing, family S member 1 |
| chr2_+_54683419 | 0.12 |
ENST00000356805.4
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr17_+_67498538 | 0.12 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr11_-_67981295 | 0.12 |
ENST00000405515.1
|
SUV420H1
|
suppressor of variegation 4-20 homolog 1 (Drosophila) |
| chr4_-_138453559 | 0.12 |
ENST00000511115.1
|
PCDH18
|
protocadherin 18 |
| chr11_+_46402583 | 0.11 |
ENST00000359803.3
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr14_-_91884150 | 0.11 |
ENST00000553403.1
|
CCDC88C
|
coiled-coil domain containing 88C |
| chrX_-_63425561 | 0.11 |
ENST00000374869.3
ENST00000330258.3 |
AMER1
|
APC membrane recruitment protein 1 |
| chr3_+_183353356 | 0.11 |
ENST00000242810.6
ENST00000493074.1 ENST00000437402.1 ENST00000454495.2 ENST00000473045.1 ENST00000468101.1 ENST00000427201.2 ENST00000482138.1 ENST00000454652.2 |
KLHL24
|
kelch-like family member 24 |
| chr15_+_89631647 | 0.11 |
ENST00000569550.1
ENST00000565066.1 ENST00000565973.1 |
ABHD2
|
abhydrolase domain containing 2 |
| chr13_-_41593425 | 0.11 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr3_+_151986709 | 0.11 |
ENST00000495875.2
ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr20_+_48884002 | 0.11 |
ENST00000425497.1
ENST00000445003.1 |
RP11-290F20.3
|
RP11-290F20.3 |
| chr1_-_93645818 | 0.11 |
ENST00000370280.1
ENST00000479918.1 |
TMED5
|
transmembrane emp24 protein transport domain containing 5 |
| chr1_-_32801825 | 0.11 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr7_+_102553430 | 0.11 |
ENST00000339431.4
ENST00000249377.4 |
LRRC17
|
leucine rich repeat containing 17 |
| chr2_+_153191706 | 0.10 |
ENST00000288670.9
|
FMNL2
|
formin-like 2 |
| chr15_+_85923856 | 0.10 |
ENST00000560302.1
ENST00000394518.2 ENST00000361243.2 ENST00000560256.1 |
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr15_-_73076030 | 0.10 |
ENST00000311669.8
|
ADPGK
|
ADP-dependent glucokinase |
| chr2_-_70475586 | 0.10 |
ENST00000416149.2
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr2_-_166060571 | 0.10 |
ENST00000360093.3
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr5_-_114515734 | 0.10 |
ENST00000514154.1
ENST00000282369.3 |
TRIM36
|
tripartite motif containing 36 |
| chr11_+_5509915 | 0.09 |
ENST00000322641.5
|
OR52D1
|
olfactory receptor, family 52, subfamily D, member 1 |
| chr1_+_25071848 | 0.09 |
ENST00000374379.4
|
CLIC4
|
chloride intracellular channel 4 |
| chr11_+_46402744 | 0.09 |
ENST00000533952.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr2_-_166060552 | 0.09 |
ENST00000283254.7
ENST00000453007.1 |
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr7_-_150924121 | 0.09 |
ENST00000441774.1
ENST00000222388.2 ENST00000287844.2 |
ABCF2
|
ATP-binding cassette, sub-family F (GCN20), member 2 |
| chr9_-_140196703 | 0.09 |
ENST00000356628.2
|
NRARP
|
NOTCH-regulated ankyrin repeat protein |
| chr1_-_26232951 | 0.09 |
ENST00000426559.2
ENST00000455785.2 |
STMN1
|
stathmin 1 |
| chr16_+_56970567 | 0.09 |
ENST00000563911.1
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr5_+_140792614 | 0.09 |
ENST00000398610.2
|
PCDHGA10
|
protocadherin gamma subfamily A, 10 |
| chr11_-_76155700 | 0.09 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr19_-_51472823 | 0.09 |
ENST00000310157.2
|
KLK6
|
kallikrein-related peptidase 6 |
| chr9_+_108463234 | 0.08 |
ENST00000374688.1
|
TMEM38B
|
transmembrane protein 38B |
| chr13_+_24144509 | 0.08 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr12_+_104359641 | 0.08 |
ENST00000537100.1
|
TDG
|
thymine-DNA glycosylase |
| chr11_-_44971702 | 0.08 |
ENST00000533940.1
ENST00000533937.1 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr3_-_18466026 | 0.08 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr3_+_14860469 | 0.08 |
ENST00000285046.5
|
FGD5
|
FYVE, RhoGEF and PH domain containing 5 |
| chr8_+_126442563 | 0.08 |
ENST00000311922.3
|
TRIB1
|
tribbles pseudokinase 1 |
| chr3_+_112930306 | 0.08 |
ENST00000495514.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr7_+_107224364 | 0.08 |
ENST00000491150.1
|
BCAP29
|
B-cell receptor-associated protein 29 |
| chr13_+_111767650 | 0.08 |
ENST00000449979.1
ENST00000370623.3 |
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr9_+_75263565 | 0.08 |
ENST00000396237.3
|
TMC1
|
transmembrane channel-like 1 |
| chr5_-_39425222 | 0.08 |
ENST00000320816.6
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr3_+_135741576 | 0.08 |
ENST00000334546.2
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr4_-_103266355 | 0.08 |
ENST00000424970.2
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr11_+_60691924 | 0.08 |
ENST00000544065.1
ENST00000453848.2 ENST00000005286.4 |
TMEM132A
|
transmembrane protein 132A |
| chr2_-_217560248 | 0.08 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr17_-_72358001 | 0.08 |
ENST00000375366.3
|
BTBD17
|
BTB (POZ) domain containing 17 |
| chr19_-_44124019 | 0.08 |
ENST00000300811.3
|
ZNF428
|
zinc finger protein 428 |
| chr4_-_186732048 | 0.07 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chrX_+_129473859 | 0.07 |
ENST00000424447.1
|
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr18_-_32924372 | 0.07 |
ENST00000261332.6
ENST00000399061.3 |
ZNF24
|
zinc finger protein 24 |
| chr6_-_127840453 | 0.07 |
ENST00000556132.1
|
SOGA3
|
SOGA family member 3 |
| chr11_+_65339820 | 0.07 |
ENST00000316409.2
ENST00000449319.2 ENST00000530349.1 |
FAM89B
|
family with sequence similarity 89, member B |
| chr17_-_39041479 | 0.07 |
ENST00000167588.3
|
KRT20
|
keratin 20 |
| chr1_-_156828810 | 0.07 |
ENST00000368195.3
|
INSRR
|
insulin receptor-related receptor |
| chr4_+_70861647 | 0.07 |
ENST00000246895.4
ENST00000381060.2 |
STATH
|
statherin |
| chr11_+_125365110 | 0.07 |
ENST00000527818.1
|
AP000708.1
|
AP000708.1 |
| chr3_+_133118839 | 0.07 |
ENST00000302334.2
|
BFSP2
|
beaded filament structural protein 2, phakinin |
| chr9_-_88896977 | 0.07 |
ENST00000311534.6
|
ISCA1
|
iron-sulfur cluster assembly 1 |
| chr1_+_240255166 | 0.07 |
ENST00000319653.9
|
FMN2
|
formin 2 |
| chr13_+_76445187 | 0.07 |
ENST00000318245.4
|
C13orf45
|
chromosome 13 open reading frame 45 |
| chr4_-_68620053 | 0.07 |
ENST00000420975.2
ENST00000226413.4 |
GNRHR
|
gonadotropin-releasing hormone receptor |
| chr19_+_10713112 | 0.07 |
ENST00000590382.1
ENST00000407327.4 |
SLC44A2
|
solute carrier family 44 (choline transporter), member 2 |
| chr12_+_104359614 | 0.07 |
ENST00000266775.9
ENST00000544861.1 |
TDG
|
thymine-DNA glycosylase |
| chr3_-_57583130 | 0.07 |
ENST00000303436.6
|
ARF4
|
ADP-ribosylation factor 4 |
| chr1_-_21503337 | 0.07 |
ENST00000400422.1
ENST00000602326.1 ENST00000411888.1 ENST00000438975.1 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr22_+_29168652 | 0.07 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chrX_+_135579670 | 0.06 |
ENST00000218364.4
|
HTATSF1
|
HIV-1 Tat specific factor 1 |
| chr16_-_29478016 | 0.06 |
ENST00000549858.1
ENST00000551411.1 |
RP11-345J4.3
|
Uncharacterized protein |
| chr4_+_113152978 | 0.06 |
ENST00000309703.6
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr12_+_64238539 | 0.06 |
ENST00000357825.3
|
SRGAP1
|
SLIT-ROBO Rho GTPase activating protein 1 |
| chr5_-_134871639 | 0.06 |
ENST00000314744.4
|
NEUROG1
|
neurogenin 1 |
| chr11_+_46402482 | 0.06 |
ENST00000441869.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr15_+_77224045 | 0.06 |
ENST00000320963.5
ENST00000394883.3 |
RCN2
|
reticulocalbin 2, EF-hand calcium binding domain |
| chr6_-_167040731 | 0.06 |
ENST00000265678.4
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr14_-_91884115 | 0.06 |
ENST00000389857.6
|
CCDC88C
|
coiled-coil domain containing 88C |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.2 | 0.6 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.1 | 0.4 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 0.3 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.1 | 0.4 | GO:0070662 | mast cell proliferation(GO:0070662) |
| 0.1 | 0.5 | GO:0035549 | positive regulation of interferon-beta secretion(GO:0035549) |
| 0.1 | 0.4 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.1 | 0.3 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.1 | 0.3 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.1 | 0.2 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.1 | 0.2 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 0.3 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.1 | 0.3 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.2 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.3 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.8 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.8 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.2 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.6 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.2 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 1.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.2 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.2 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.3 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.7 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.4 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.2 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.6 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.1 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.1 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.0 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.1 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.0 | GO:0034241 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.1 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.0 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.0 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.4 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.3 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:1990015 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.0 | 1.0 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.3 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.2 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.0 | GO:0034680 | integrin alpha1-beta1 complex(GO:0034665) integrin alpha3-beta1 complex(GO:0034667) integrin alpha10-beta1 complex(GO:0034680) integrin alpha11-beta1 complex(GO:0034681) |
| 0.0 | 0.2 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0033677 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) DNA/RNA helicase activity(GO:0033677) |
| 0.1 | 0.3 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.1 | 0.6 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 1.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.2 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.2 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.2 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.3 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.2 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.3 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 1.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.1 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |