Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
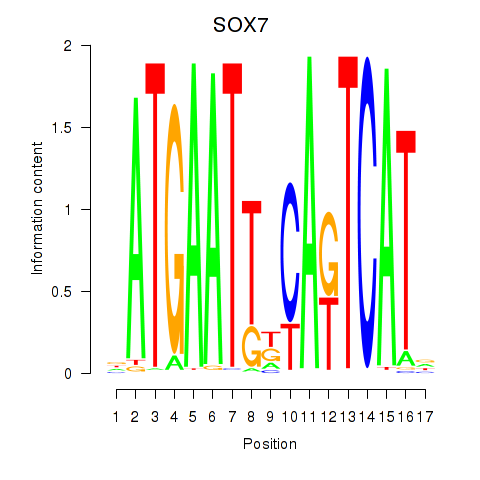
Results for SOX7
Z-value: 0.52
Transcription factors associated with SOX7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX7
|
ENSG00000171056.6 | SRY-box transcription factor 7 |
|
SOX7
|
ENSG00000258724.1 | SRY-box transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX7 | hg19_v2_chr8_-_10697281_10697365 | -0.39 | 5.2e-02 | Click! |
Activity profile of SOX7 motif
Sorted Z-values of SOX7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_190445499 | 1.82 |
ENST00000261024.2
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr17_-_66951474 | 1.42 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr19_-_38916802 | 1.26 |
ENST00000587738.1
|
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr19_-_38916839 | 1.06 |
ENST00000433821.2
ENST00000426920.2 ENST00000587753.1 ENST00000454404.2 ENST00000293062.9 |
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr5_-_95158644 | 1.04 |
ENST00000237858.6
|
GLRX
|
glutaredoxin (thioltransferase) |
| chr19_-_38916822 | 1.02 |
ENST00000586305.1
|
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr11_+_7618413 | 1.02 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr5_-_95158375 | 0.97 |
ENST00000512469.2
ENST00000379979.4 ENST00000505427.1 ENST00000508780.1 |
GLRX
|
glutaredoxin (thioltransferase) |
| chr3_+_107318157 | 0.85 |
ENST00000406780.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr6_-_111804905 | 0.70 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr12_-_90049878 | 0.59 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr13_-_37633567 | 0.57 |
ENST00000464744.1
|
SUPT20H
|
suppressor of Ty 20 homolog (S. cerevisiae) |
| chr12_-_90049828 | 0.57 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr12_-_57443886 | 0.56 |
ENST00000300119.3
|
MYO1A
|
myosin IA |
| chr16_-_28303360 | 0.46 |
ENST00000501520.1
|
RP11-57A19.2
|
RP11-57A19.2 |
| chr9_-_70465758 | 0.45 |
ENST00000489273.1
|
CBWD5
|
COBW domain containing 5 |
| chr19_+_37837185 | 0.44 |
ENST00000541583.2
|
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr8_-_19459993 | 0.44 |
ENST00000454498.2
ENST00000520003.1 |
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr2_+_179317994 | 0.43 |
ENST00000375129.4
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr18_+_32820990 | 0.41 |
ENST00000601719.1
ENST00000591206.1 ENST00000330501.7 ENST00000261333.6 ENST00000355632.4 ENST00000585800.1 |
ZNF397
|
zinc finger protein 397 |
| chr13_-_28024681 | 0.39 |
ENST00000381116.1
ENST00000381120.3 ENST00000431572.2 |
MTIF3
|
mitochondrial translational initiation factor 3 |
| chr17_-_56082455 | 0.36 |
ENST00000578794.1
|
RP11-159D12.5
|
Uncharacterized protein |
| chr6_-_29324054 | 0.36 |
ENST00000543825.1
|
OR5V1
|
olfactory receptor, family 5, subfamily V, member 1 |
| chrX_+_69353284 | 0.35 |
ENST00000342206.6
ENST00000356413.4 |
IGBP1
|
immunoglobulin (CD79A) binding protein 1 |
| chr6_+_147527103 | 0.31 |
ENST00000179882.6
|
STXBP5
|
syntaxin binding protein 5 (tomosyn) |
| chr11_-_62474803 | 0.31 |
ENST00000533982.1
ENST00000360796.5 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr6_-_3195981 | 0.31 |
ENST00000425384.2
ENST00000435043.2 |
RP1-40E16.9
|
RP1-40E16.9 |
| chrY_+_26997726 | 0.30 |
ENST00000382296.2
|
DAZ4
|
deleted in azoospermia 4 |
| chr12_-_58027138 | 0.29 |
ENST00000341156.4
|
B4GALNT1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
| chr1_+_100598691 | 0.26 |
ENST00000370143.1
ENST00000370141.2 |
TRMT13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr10_+_5005445 | 0.26 |
ENST00000380872.4
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr6_-_86303833 | 0.25 |
ENST00000505648.1
|
SNX14
|
sorting nexin 14 |
| chr10_-_5046042 | 0.24 |
ENST00000421196.3
ENST00000455190.1 |
AKR1C2
|
aldo-keto reductase family 1, member C2 |
| chr8_-_102803163 | 0.23 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr10_-_12084770 | 0.22 |
ENST00000357604.5
|
UPF2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr6_+_80129989 | 0.20 |
ENST00000429444.1
|
RP1-232L24.3
|
RP1-232L24.3 |
| chr6_+_144164455 | 0.20 |
ENST00000367576.5
|
LTV1
|
LTV1 homolog (S. cerevisiae) |
| chr12_+_64173583 | 0.19 |
ENST00000261234.6
|
TMEM5
|
transmembrane protein 5 |
| chr14_-_102771462 | 0.19 |
ENST00000522874.1
|
MOK
|
MOK protein kinase |
| chr4_+_69313145 | 0.18 |
ENST00000305363.4
|
TMPRSS11E
|
transmembrane protease, serine 11E |
| chr16_+_28303804 | 0.18 |
ENST00000341901.4
|
SBK1
|
SH3 domain binding kinase 1 |
| chr1_+_223101757 | 0.17 |
ENST00000284476.6
|
DISP1
|
dispatched homolog 1 (Drosophila) |
| chr21_-_15918618 | 0.15 |
ENST00000400564.1
ENST00000400566.1 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr15_+_25068773 | 0.14 |
ENST00000400100.1
ENST00000400098.1 |
SNRPN
|
small nuclear ribonucleoprotein polypeptide N |
| chr1_+_199996733 | 0.12 |
ENST00000236914.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr19_-_21950332 | 0.11 |
ENST00000598026.1
|
ZNF100
|
zinc finger protein 100 |
| chr4_-_177190364 | 0.11 |
ENST00000296525.3
|
ASB5
|
ankyrin repeat and SOCS box containing 5 |
| chr5_+_175511859 | 0.11 |
ENST00000503724.2
ENST00000253490.4 |
FAM153B
|
family with sequence similarity 153, member B |
| chr14_-_106573756 | 0.10 |
ENST00000390601.2
|
IGHV3-11
|
immunoglobulin heavy variable 3-11 (gene/pseudogene) |
| chr19_-_22379753 | 0.10 |
ENST00000397121.2
|
ZNF676
|
zinc finger protein 676 |
| chr6_+_150690133 | 0.09 |
ENST00000392255.3
ENST00000500320.3 |
IYD
|
iodotyrosine deiodinase |
| chr10_-_17243579 | 0.08 |
ENST00000525762.1
ENST00000412821.3 ENST00000351358.4 ENST00000377766.5 ENST00000358282.7 ENST00000488990.1 ENST00000377799.3 |
TRDMT1
|
tRNA aspartic acid methyltransferase 1 |
| chr2_-_166930131 | 0.07 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr22_+_23101182 | 0.06 |
ENST00000390312.2
|
IGLV2-14
|
immunoglobulin lambda variable 2-14 |
| chr6_-_132910877 | 0.06 |
ENST00000258034.2
|
TAAR5
|
trace amine associated receptor 5 |
| chr16_-_279405 | 0.05 |
ENST00000430864.1
ENST00000293872.8 ENST00000337351.4 ENST00000397783.1 |
LUC7L
|
LUC7-like (S. cerevisiae) |
| chr11_+_55944094 | 0.05 |
ENST00000312298.1
|
OR5J2
|
olfactory receptor, family 5, subfamily J, member 2 |
| chr15_+_59439899 | 0.05 |
ENST00000599727.1
|
C15ORF31
|
C15ORF31 |
| chr1_-_186430222 | 0.05 |
ENST00000391997.2
|
PDC
|
phosducin |
| chr5_+_161494521 | 0.04 |
ENST00000356592.3
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr1_+_10509971 | 0.03 |
ENST00000320498.4
|
CORT
|
cortistatin |
| chr12_-_57328187 | 0.03 |
ENST00000293502.1
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr1_-_54303934 | 0.03 |
ENST00000537333.1
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr6_+_150690028 | 0.03 |
ENST00000229447.5
ENST00000344419.3 |
IYD
|
iodotyrosine deiodinase |
| chr1_+_158901329 | 0.02 |
ENST00000368140.1
ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1
|
pyrin and HIN domain family, member 1 |
| chr21_-_21630994 | 0.02 |
ENST00000436373.1
|
AP001171.1
|
AP001171.1 |
| chr21_+_34144411 | 0.02 |
ENST00000382375.4
ENST00000453404.1 ENST00000382378.1 ENST00000477513.1 |
C21orf49
|
chromosome 21 open reading frame 49 |
| chr16_+_33020496 | 0.01 |
ENST00000565407.2
|
IGHV3OR16-8
|
immunoglobulin heavy variable 3/OR16-8 (non-functional) |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 0.1 | 1.2 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.1 | 1.4 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 0.4 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.1 | 2.0 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.1 | 0.4 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.1 | 3.3 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.1 | 0.5 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.7 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.6 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.4 | GO:0060632 | regulation of microtubule-based movement(GO:0060632) |
| 0.0 | 0.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.3 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 1.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.6 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0034448 | EGO complex(GO:0034448) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.0 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.2 | 1.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.2 | 2.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 0.4 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.1 | 3.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.3 | GO:0003947 | (N-acetylneuraminyl)-galactosylglucosylceramide N-acetylgalactosaminyltransferase activity(GO:0003947) |
| 0.1 | 0.5 | GO:0018636 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) |
| 0.0 | 1.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | PID RAS PATHWAY | Regulation of Ras family activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.8 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |