Project
Inflammatory response time course, HUVEC (Wada et al., 2009)
Navigation
Downloads
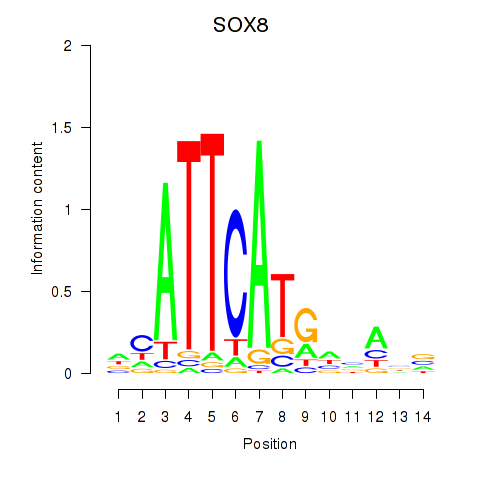
Results for SOX8
Z-value: 0.75
Transcription factors associated with SOX8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX8
|
ENSG00000005513.9 | SRY-box transcription factor 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX8 | hg19_v2_chr16_+_1031762_1031808 | 0.11 | 6.1e-01 | Click! |
Activity profile of SOX8 motif
Sorted Z-values of SOX8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_47352477 | 1.45 |
ENST00000593412.1
|
PRED62
|
Uncharacterized protein |
| chr2_-_175711133 | 1.23 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr15_+_45926919 | 0.89 |
ENST00000561735.1
ENST00000260324.7 |
SQRDL
|
sulfide quinone reductase-like (yeast) |
| chr4_-_47983519 | 0.86 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr10_+_88414338 | 0.84 |
ENST00000241891.5
ENST00000443292.1 |
OPN4
|
opsin 4 |
| chr10_+_88414298 | 0.83 |
ENST00000372071.2
|
OPN4
|
opsin 4 |
| chr10_+_103986085 | 0.79 |
ENST00000370005.3
|
ELOVL3
|
ELOVL fatty acid elongase 3 |
| chr1_+_13516066 | 0.73 |
ENST00000332192.6
|
PRAMEF21
|
PRAME family member 21 |
| chr20_-_44176013 | 0.69 |
ENST00000555685.1
|
EPPIN
|
epididymal peptidase inhibitor |
| chr7_+_100303676 | 0.67 |
ENST00000303151.4
|
POP7
|
processing of precursor 7, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr20_-_5426332 | 0.66 |
ENST00000420529.1
|
LINC00658
|
long intergenic non-protein coding RNA 658 |
| chr2_+_233527443 | 0.59 |
ENST00000410095.1
|
EFHD1
|
EF-hand domain family, member D1 |
| chr15_-_58306295 | 0.56 |
ENST00000559517.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr12_-_39837192 | 0.56 |
ENST00000361961.3
ENST00000395670.3 |
KIF21A
|
kinesin family member 21A |
| chr3_-_52090461 | 0.53 |
ENST00000296483.6
ENST00000495880.1 |
DUSP7
|
dual specificity phosphatase 7 |
| chr5_-_146781153 | 0.53 |
ENST00000520473.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr4_+_86525299 | 0.52 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr4_+_74347400 | 0.52 |
ENST00000226355.3
|
AFM
|
afamin |
| chr12_-_10151773 | 0.51 |
ENST00000298527.6
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1, member B |
| chr18_-_33702078 | 0.50 |
ENST00000586829.1
|
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chrX_+_18725758 | 0.48 |
ENST00000472826.1
ENST00000544635.1 ENST00000496075.2 |
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr4_+_89299994 | 0.48 |
ENST00000264346.7
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr17_-_34207295 | 0.45 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr14_+_75745477 | 0.44 |
ENST00000303562.4
ENST00000554617.1 ENST00000554212.1 ENST00000535987.1 ENST00000555242.1 |
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr3_-_123339418 | 0.43 |
ENST00000583087.1
|
MYLK
|
myosin light chain kinase |
| chr15_+_34261089 | 0.42 |
ENST00000383263.5
|
CHRM5
|
cholinergic receptor, muscarinic 5 |
| chr2_+_89986318 | 0.41 |
ENST00000491977.1
|
IGKV2D-29
|
immunoglobulin kappa variable 2D-29 |
| chr17_+_41132564 | 0.39 |
ENST00000361677.1
ENST00000589705.1 |
RUNDC1
|
RUN domain containing 1 |
| chr19_-_37663332 | 0.39 |
ENST00000392157.2
|
ZNF585A
|
zinc finger protein 585A |
| chr16_+_57392684 | 0.38 |
ENST00000219235.4
|
CCL22
|
chemokine (C-C motif) ligand 22 |
| chr3_-_54962100 | 0.38 |
ENST00000273286.5
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1 |
| chr1_+_26496362 | 0.38 |
ENST00000374266.5
ENST00000270812.5 |
ZNF593
|
zinc finger protein 593 |
| chr12_+_45686457 | 0.37 |
ENST00000441606.2
|
ANO6
|
anoctamin 6 |
| chr12_-_123201337 | 0.37 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr1_+_13742808 | 0.36 |
ENST00000602960.1
|
PRAMEF20
|
PRAME family member 20 |
| chr18_+_61575200 | 0.36 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr1_+_110577229 | 0.36 |
ENST00000369795.3
ENST00000369794.2 |
STRIP1
|
striatin interacting protein 1 |
| chr14_-_21562648 | 0.36 |
ENST00000555270.1
|
ZNF219
|
zinc finger protein 219 |
| chr4_+_156680153 | 0.35 |
ENST00000502959.1
ENST00000505764.1 ENST00000507146.1 ENST00000264424.8 ENST00000503520.1 |
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr1_-_200992827 | 0.34 |
ENST00000332129.2
ENST00000422435.2 |
KIF21B
|
kinesin family member 21B |
| chr3_-_47934234 | 0.34 |
ENST00000420772.2
|
MAP4
|
microtubule-associated protein 4 |
| chr2_+_74425689 | 0.33 |
ENST00000394053.2
ENST00000409804.1 ENST00000264090.4 ENST00000394050.3 ENST00000409601.1 |
MTHFD2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr7_-_122339162 | 0.32 |
ENST00000340112.2
|
RNF133
|
ring finger protein 133 |
| chr5_-_160279207 | 0.32 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr18_+_32173276 | 0.32 |
ENST00000591816.1
ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA
|
dystrobrevin, alpha |
| chr2_-_120124258 | 0.32 |
ENST00000409877.1
ENST00000409523.1 ENST00000409466.2 ENST00000414534.1 |
C2orf76
|
chromosome 2 open reading frame 76 |
| chr1_-_222763240 | 0.31 |
ENST00000352967.4
ENST00000391882.1 ENST00000543857.1 |
TAF1A
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa |
| chr3_+_173302222 | 0.31 |
ENST00000361589.4
|
NLGN1
|
neuroligin 1 |
| chr5_-_94417339 | 0.31 |
ENST00000429576.2
ENST00000508509.1 ENST00000510732.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr4_+_156680143 | 0.31 |
ENST00000505154.1
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr19_+_45681997 | 0.31 |
ENST00000433642.2
|
BLOC1S3
|
biogenesis of lysosomal organelles complex-1, subunit 3 |
| chr11_+_10477733 | 0.31 |
ENST00000528723.1
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr15_-_89755034 | 0.31 |
ENST00000563254.1
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr1_-_231005310 | 0.31 |
ENST00000470540.1
|
C1orf198
|
chromosome 1 open reading frame 198 |
| chr12_+_113354341 | 0.31 |
ENST00000553152.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr12_-_50101165 | 0.31 |
ENST00000352151.5
ENST00000335154.5 ENST00000293590.5 |
FMNL3
|
formin-like 3 |
| chr2_-_163008903 | 0.30 |
ENST00000418842.2
ENST00000375497.3 |
GCG
|
glucagon |
| chr4_-_69536346 | 0.30 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr6_-_136788001 | 0.29 |
ENST00000544465.1
|
MAP7
|
microtubule-associated protein 7 |
| chr7_-_27169801 | 0.28 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr6_-_52668605 | 0.28 |
ENST00000334575.5
|
GSTA1
|
glutathione S-transferase alpha 1 |
| chr12_+_122242597 | 0.28 |
ENST00000267197.5
|
SETD1B
|
SET domain containing 1B |
| chr19_-_37663572 | 0.27 |
ENST00000588354.1
ENST00000292841.5 ENST00000355533.2 ENST00000356958.4 |
ZNF585A
|
zinc finger protein 585A |
| chr11_+_67219867 | 0.27 |
ENST00000438189.2
|
CABP4
|
calcium binding protein 4 |
| chr2_-_3606206 | 0.27 |
ENST00000315212.3
|
RNASEH1
|
ribonuclease H1 |
| chr3_-_21792838 | 0.27 |
ENST00000281523.2
|
ZNF385D
|
zinc finger protein 385D |
| chr11_+_5474638 | 0.26 |
ENST00000341449.2
|
OR51I2
|
olfactory receptor, family 51, subfamily I, member 2 |
| chr9_-_128246769 | 0.26 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr15_-_50557863 | 0.26 |
ENST00000543581.1
|
HDC
|
histidine decarboxylase |
| chr4_+_155484155 | 0.26 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr20_+_48807351 | 0.26 |
ENST00000303004.3
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr2_+_26624775 | 0.25 |
ENST00000288710.2
|
DRC1
|
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
| chr11_+_57435219 | 0.25 |
ENST00000527985.1
ENST00000287169.3 |
ZDHHC5
|
zinc finger, DHHC-type containing 5 |
| chr2_+_97203082 | 0.25 |
ENST00000454558.2
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr4_-_140544386 | 0.24 |
ENST00000561977.1
|
RP11-308D13.3
|
RP11-308D13.3 |
| chr11_+_12308447 | 0.24 |
ENST00000256186.2
|
MICALCL
|
MICAL C-terminal like |
| chr3_+_137906109 | 0.24 |
ENST00000481646.1
ENST00000469044.1 ENST00000491704.1 ENST00000461600.1 |
ARMC8
|
armadillo repeat containing 8 |
| chr19_+_51728316 | 0.24 |
ENST00000436584.2
ENST00000421133.2 ENST00000391796.3 ENST00000262262.4 |
CD33
|
CD33 molecule |
| chr18_+_32455201 | 0.24 |
ENST00000590831.2
|
DTNA
|
dystrobrevin, alpha |
| chr5_-_94417314 | 0.24 |
ENST00000505208.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr20_+_32399093 | 0.24 |
ENST00000217402.2
|
CHMP4B
|
charged multivesicular body protein 4B |
| chr14_+_95078714 | 0.23 |
ENST00000393078.3
ENST00000393080.4 ENST00000467132.1 |
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr3_+_173116225 | 0.23 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr2_-_223520770 | 0.23 |
ENST00000536361.1
|
FARSB
|
phenylalanyl-tRNA synthetase, beta subunit |
| chrX_-_54824673 | 0.23 |
ENST00000218436.6
|
ITIH6
|
inter-alpha-trypsin inhibitor heavy chain family, member 6 |
| chr12_-_50101003 | 0.22 |
ENST00000550488.1
|
FMNL3
|
formin-like 3 |
| chr15_-_62937380 | 0.22 |
ENST00000560347.1
ENST00000558940.1 |
RP11-625H11.1
|
Uncharacterized protein |
| chr3_+_97887544 | 0.22 |
ENST00000356526.2
|
OR5H15
|
olfactory receptor, family 5, subfamily H, member 15 |
| chr5_+_32788945 | 0.22 |
ENST00000326958.1
|
AC026703.1
|
AC026703.1 |
| chr2_+_166326157 | 0.22 |
ENST00000421875.1
ENST00000314499.7 ENST00000409664.1 |
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr9_-_113761720 | 0.22 |
ENST00000541779.1
ENST00000374430.2 |
LPAR1
|
lysophosphatidic acid receptor 1 |
| chr11_-_121986923 | 0.22 |
ENST00000560104.1
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr11_-_128457446 | 0.22 |
ENST00000392668.4
|
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr22_-_30722912 | 0.22 |
ENST00000215790.7
|
TBC1D10A
|
TBC1 domain family, member 10A |
| chr11_-_46142615 | 0.21 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chr9_-_116837249 | 0.21 |
ENST00000466610.2
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr9_+_127624387 | 0.21 |
ENST00000353214.2
|
ARPC5L
|
actin related protein 2/3 complex, subunit 5-like |
| chr12_+_20848282 | 0.21 |
ENST00000545604.1
|
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr22_-_30722866 | 0.21 |
ENST00000403477.3
|
TBC1D10A
|
TBC1 domain family, member 10A |
| chr4_-_70518941 | 0.21 |
ENST00000286604.4
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr16_+_21244986 | 0.21 |
ENST00000311620.5
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr14_+_39583427 | 0.21 |
ENST00000308317.6
ENST00000396249.2 ENST00000250379.8 ENST00000534684.2 ENST00000527381.1 |
GEMIN2
|
gem (nuclear organelle) associated protein 2 |
| chr10_-_38265517 | 0.20 |
ENST00000302609.7
|
ZNF25
|
zinc finger protein 25 |
| chr15_+_78830023 | 0.20 |
ENST00000599596.1
|
AC027228.1
|
HCG2005369; Uncharacterized protein |
| chr12_+_20848377 | 0.20 |
ENST00000540354.1
ENST00000266509.2 ENST00000381552.1 |
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr11_-_41481135 | 0.20 |
ENST00000528697.1
ENST00000530763.1 |
LRRC4C
|
leucine rich repeat containing 4C |
| chr17_+_2240916 | 0.20 |
ENST00000574563.1
|
SGSM2
|
small G protein signaling modulator 2 |
| chr16_+_87985029 | 0.20 |
ENST00000439677.1
ENST00000286122.7 ENST00000355163.5 ENST00000454563.1 ENST00000479780.2 ENST00000393208.2 ENST00000412691.1 ENST00000355022.4 |
BANP
|
BTG3 associated nuclear protein |
| chr14_+_88471468 | 0.20 |
ENST00000267549.3
|
GPR65
|
G protein-coupled receptor 65 |
| chr7_+_23145366 | 0.20 |
ENST00000339077.5
ENST00000322275.5 ENST00000539124.1 ENST00000542558.1 |
KLHL7
|
kelch-like family member 7 |
| chr3_+_14693247 | 0.20 |
ENST00000383794.3
ENST00000303688.7 |
CCDC174
|
coiled-coil domain containing 174 |
| chr19_-_8213753 | 0.19 |
ENST00000601739.1
|
FBN3
|
fibrillin 3 |
| chr3_-_55001115 | 0.19 |
ENST00000493075.1
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1 |
| chr16_-_18911366 | 0.19 |
ENST00000565224.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr7_+_64126535 | 0.18 |
ENST00000344930.3
|
ZNF107
|
zinc finger protein 107 |
| chr11_+_9482551 | 0.18 |
ENST00000438144.2
ENST00000526657.1 ENST00000299606.2 ENST00000534265.1 ENST00000412390.2 |
ZNF143
|
zinc finger protein 143 |
| chr17_-_19648683 | 0.18 |
ENST00000573368.1
ENST00000457500.2 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr4_-_100356551 | 0.18 |
ENST00000209665.4
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr3_+_577893 | 0.18 |
ENST00000420823.1
ENST00000442809.1 |
AC090044.1
|
AC090044.1 |
| chr19_+_52076425 | 0.18 |
ENST00000436511.2
|
ZNF175
|
zinc finger protein 175 |
| chr3_+_138340049 | 0.18 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr3_-_193272588 | 0.18 |
ENST00000295548.3
|
ATP13A4
|
ATPase type 13A4 |
| chr12_-_123187890 | 0.18 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr12_+_14927270 | 0.18 |
ENST00000544848.1
|
H2AFJ
|
H2A histone family, member J |
| chr1_+_144811943 | 0.18 |
ENST00000281815.8
|
NBPF9
|
neuroblastoma breakpoint family, member 9 |
| chr4_+_71248795 | 0.18 |
ENST00000304915.3
|
SMR3B
|
submaxillary gland androgen regulated protein 3B |
| chr3_+_185080908 | 0.17 |
ENST00000265026.3
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr16_+_87636474 | 0.17 |
ENST00000284262.2
|
JPH3
|
junctophilin 3 |
| chr15_-_38519066 | 0.17 |
ENST00000561320.1
ENST00000561161.1 |
RP11-346D14.1
|
RP11-346D14.1 |
| chr2_+_102413726 | 0.17 |
ENST00000350878.4
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr4_+_155484103 | 0.17 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr11_+_9482512 | 0.17 |
ENST00000396602.2
ENST00000530463.1 ENST00000533542.1 ENST00000532577.1 ENST00000396597.3 |
ZNF143
|
zinc finger protein 143 |
| chr10_-_75676400 | 0.16 |
ENST00000412307.2
|
C10orf55
|
chromosome 10 open reading frame 55 |
| chr11_+_43702322 | 0.16 |
ENST00000395700.4
|
HSD17B12
|
hydroxysteroid (17-beta) dehydrogenase 12 |
| chr6_-_31689456 | 0.16 |
ENST00000495859.1
ENST00000375819.2 |
LY6G6C
|
lymphocyte antigen 6 complex, locus G6C |
| chr8_+_7693993 | 0.16 |
ENST00000314265.2
|
DEFB104A
|
defensin, beta 104A |
| chr2_-_151344172 | 0.16 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr3_-_190167571 | 0.16 |
ENST00000354905.2
|
TMEM207
|
transmembrane protein 207 |
| chr6_+_31021225 | 0.16 |
ENST00000565192.1
ENST00000562344.1 |
HCG22
|
HLA complex group 22 |
| chr11_-_57177586 | 0.15 |
ENST00000529411.1
|
RP11-872D17.8
|
Uncharacterized protein |
| chr7_-_127255982 | 0.15 |
ENST00000338516.3
ENST00000378740.2 |
PAX4
|
paired box 4 |
| chr11_-_62995986 | 0.15 |
ENST00000403374.2
|
SLC22A25
|
solute carrier family 22, member 25 |
| chr6_-_138539627 | 0.15 |
ENST00000527246.2
|
PBOV1
|
prostate and breast cancer overexpressed 1 |
| chr8_+_38758737 | 0.15 |
ENST00000521746.1
ENST00000420274.1 |
PLEKHA2
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 |
| chr19_+_1524072 | 0.15 |
ENST00000454744.2
|
PLK5
|
polo-like kinase 5 |
| chr17_-_8021710 | 0.15 |
ENST00000380149.1
ENST00000448843.2 |
ALOXE3
|
arachidonate lipoxygenase 3 |
| chr10_-_4285923 | 0.15 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr16_+_618837 | 0.15 |
ENST00000409439.2
|
PIGQ
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr14_-_106642049 | 0.15 |
ENST00000390605.2
|
IGHV1-18
|
immunoglobulin heavy variable 1-18 |
| chr8_-_20040638 | 0.15 |
ENST00000519026.1
ENST00000276373.5 ENST00000440926.1 ENST00000437980.1 |
SLC18A1
|
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr16_+_53241854 | 0.14 |
ENST00000565803.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr11_+_60995849 | 0.14 |
ENST00000537932.1
|
PGA4
|
pepsinogen 4, group I (pepsinogen A) |
| chr12_+_82347498 | 0.14 |
ENST00000550506.1
|
RP11-362A1.1
|
RP11-362A1.1 |
| chr8_-_91095099 | 0.14 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr7_-_38289173 | 0.14 |
ENST00000436911.2
|
TRGC2
|
T cell receptor gamma constant 2 |
| chr16_+_31271274 | 0.14 |
ENST00000287497.8
ENST00000544665.3 |
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit) |
| chr7_+_80275752 | 0.14 |
ENST00000419819.2
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr22_-_50708781 | 0.14 |
ENST00000449719.2
ENST00000330651.6 |
MAPK11
|
mitogen-activated protein kinase 11 |
| chr12_+_133066137 | 0.14 |
ENST00000434748.2
|
FBRSL1
|
fibrosin-like 1 |
| chr9_-_130533615 | 0.14 |
ENST00000373277.4
|
SH2D3C
|
SH2 domain containing 3C |
| chr15_+_62853562 | 0.13 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr3_+_156008809 | 0.13 |
ENST00000302490.8
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr18_-_47376197 | 0.13 |
ENST00000592688.1
|
MYO5B
|
myosin VB |
| chr14_+_52327350 | 0.13 |
ENST00000555472.1
ENST00000556766.1 |
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr11_+_112130988 | 0.13 |
ENST00000595053.1
|
AP002884.2
|
LOC100132686 protein; Uncharacterized protein |
| chr16_+_30075783 | 0.13 |
ENST00000412304.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr16_+_30075463 | 0.13 |
ENST00000562168.1
ENST00000569545.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr7_+_23145347 | 0.13 |
ENST00000322231.7
|
KLHL7
|
kelch-like family member 7 |
| chr16_+_30075595 | 0.12 |
ENST00000563060.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr7_-_229557 | 0.12 |
ENST00000514988.1
|
AC145676.2
|
Uncharacterized protein |
| chr5_-_160973649 | 0.12 |
ENST00000393959.1
ENST00000517547.1 |
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr11_+_125774362 | 0.12 |
ENST00000530414.1
ENST00000530129.2 |
DDX25
|
DEAD (Asp-Glu-Ala-Asp) box helicase 25 |
| chr1_-_238649319 | 0.12 |
ENST00000400946.2
|
RP11-371I1.2
|
long intergenic non-protein coding RNA 1139 |
| chr2_+_158114051 | 0.12 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr18_-_5396271 | 0.12 |
ENST00000579951.1
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr3_-_186288097 | 0.12 |
ENST00000446782.1
|
TBCCD1
|
TBCC domain containing 1 |
| chr19_-_47287990 | 0.12 |
ENST00000593713.1
ENST00000598022.1 ENST00000434726.2 |
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr17_-_66597530 | 0.11 |
ENST00000592554.1
|
FAM20A
|
family with sequence similarity 20, member A |
| chr4_-_38858428 | 0.11 |
ENST00000436693.2
ENST00000508254.1 ENST00000514655.1 ENST00000506146.1 |
TLR6
TLR1
|
toll-like receptor 6 toll-like receptor 1 |
| chr11_+_74862140 | 0.11 |
ENST00000525650.1
ENST00000454962.2 |
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1 |
| chr6_+_32605134 | 0.11 |
ENST00000343139.5
ENST00000395363.1 ENST00000496318.1 |
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr9_-_95244781 | 0.11 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr20_+_32150140 | 0.11 |
ENST00000344201.3
ENST00000346541.3 ENST00000397800.1 ENST00000397798.2 ENST00000492345.1 |
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr1_+_153330322 | 0.11 |
ENST00000368738.3
|
S100A9
|
S100 calcium binding protein A9 |
| chr1_+_152956549 | 0.10 |
ENST00000307122.2
|
SPRR1A
|
small proline-rich protein 1A |
| chr17_-_41132410 | 0.10 |
ENST00000409446.3
ENST00000453594.1 ENST00000409399.1 ENST00000421990.2 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr3_+_98216448 | 0.10 |
ENST00000427338.1
|
OR5K2
|
olfactory receptor, family 5, subfamily K, member 2 |
| chr22_-_43411106 | 0.10 |
ENST00000453643.1
ENST00000263246.3 ENST00000337959.4 |
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr4_-_10686373 | 0.10 |
ENST00000442825.2
|
CLNK
|
cytokine-dependent hematopoietic cell linker |
| chr18_-_3845321 | 0.10 |
ENST00000539435.1
ENST00000400147.2 |
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr12_-_11548496 | 0.10 |
ENST00000389362.4
ENST00000565533.1 ENST00000546254.1 |
PRB2
PRB1
|
proline-rich protein BstNI subfamily 2 proline-rich protein BstNI subfamily 1 |
| chr6_-_116833500 | 0.10 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr12_-_100660833 | 0.10 |
ENST00000551642.1
ENST00000416321.1 ENST00000550587.1 ENST00000549249.1 |
DEPDC4
|
DEP domain containing 4 |
| chrX_+_15767971 | 0.10 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr9_+_84603687 | 0.09 |
ENST00000344803.2
|
SPATA31D1
|
SPATA31 subfamily D, member 1 |
| chr3_+_35683651 | 0.09 |
ENST00000187397.4
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr1_-_247921982 | 0.09 |
ENST00000408896.2
|
OR1C1
|
olfactory receptor, family 1, subfamily C, member 1 |
| chr1_-_38512450 | 0.09 |
ENST00000373012.2
|
POU3F1
|
POU class 3 homeobox 1 |
| chr22_-_50523760 | 0.09 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr5_-_78809950 | 0.09 |
ENST00000334082.6
|
HOMER1
|
homer homolog 1 (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0099545 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.1 | 0.5 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.7 | GO:1901318 | negative regulation of sperm motility(GO:1901318) |
| 0.1 | 1.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.4 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.1 | 0.3 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.8 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.3 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.1 | 0.7 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 | 0.3 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 0.2 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.1 | 0.6 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.1 | 0.4 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 0.2 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.4 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.2 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 0.3 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.2 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.4 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.3 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.4 | GO:0071726 | toll-like receptor TLR6:TLR2 signaling pathway(GO:0038124) response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.0 | 0.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.3 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 1.3 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.6 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.2 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.0 | 0.1 | GO:1903803 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 1.2 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.5 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.2 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.4 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.3 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.0 | 0.5 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:1901382 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.1 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.0 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.1 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.3 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.0 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.6 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.1 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.5 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.3 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.1 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.8 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.9 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.7 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.4 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 1.7 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.3 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.1 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.1 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 0.5 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.4 | GO:0000035 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.3 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 0.8 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.9 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.5 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.7 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.2 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.0 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.5 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.0 | 0.6 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 1.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.5 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.0 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.4 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.0 | 0.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |